6LO8
 
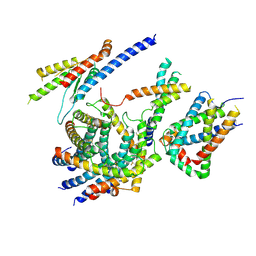 | | Cryo-EM structure of the TIM22 complex from yeast | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM10, Mitochondrial import inner membrane translocase subunit TIM12, Mitochondrial import inner membrane translocase subunit TIM18, ... | | Authors: | Zhang, Y, Zhou, X, Wu, X, Li, L. | | Deposit date: | 2020-01-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the mitochondrial TIM22 complex from yeast.
Cell Res., 31, 2021
|
|
3MXX
 
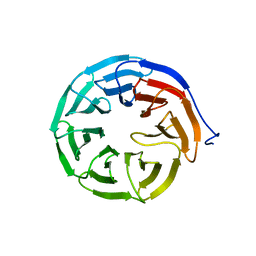 | |
3N0D
 
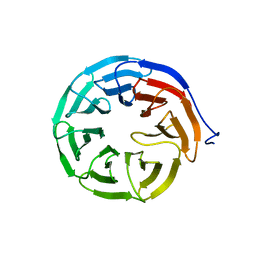 | |
3N0E
 
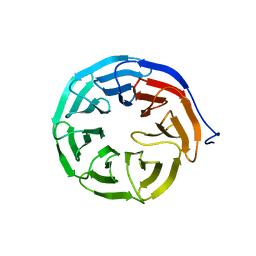 | |
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9J1T
 
 | | Structure of a triple-helix region of human Collagen type IV from Trautec | | Descriptor: | GLYCEROL, Triple-helix region of human collagen type IV | | Authors: | Fan, X, Chu, Y, Zhai, Y, Fu, S, Li, D, Feng, P, Cao, K, Wu, X, Cai, H, Wang, H, Qian, S. | | Deposit date: | 2024-08-05 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type IV from Trautec
To Be Published
|
|
9J26
 
 | | Structure of a triple-helix region of human Collagen type IV from Trautec | | Descriptor: | Triple-helix region of human collagen type IV | | Authors: | Fan, X, Zhai, Y, Chu, Y, Fu, S, Li, D, Cao, K, Feng, P, Wu, X, Cai, H, Ma, L, Qian, S. | | Deposit date: | 2024-08-06 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type IV from Trautec
To Be Published
|
|
8EFF
 
 | | CryoEM of the soluble OPA1 tetramer from the GDP-AlFx bound helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EEW
 
 | | CryoEM of the soluble OPA1 dimer from the GDP-AlFx bound helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFS
 
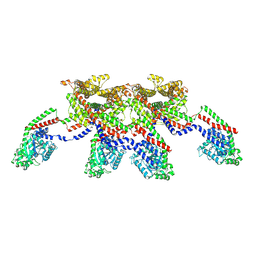 | | CryoEM of the soluble OPA1 tetramer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EF7
 
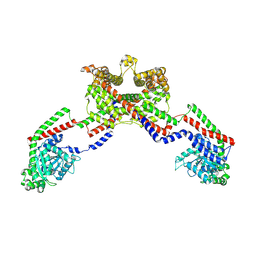 | | CryoEM of the soluble OPA1 dimer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
8EFT
 
 | | CryoEM of the soluble OPA1 interfaces from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
7XX4
 
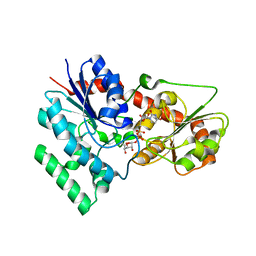 | | designed glycosyltransferase | | Descriptor: | Oleandomycin glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Lu, M, Wu, X. | | Deposit date: | 2022-05-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Design of a chimeric glycosyltransferase OleD for the site-specific O-monoglycosylation of 3-hydroxypyridine in nosiheptide.
Microb Biotechnol, 16, 2023
|
|
6PLM
 
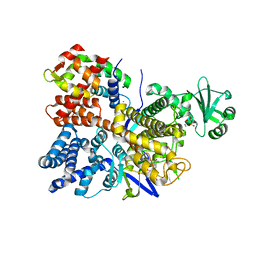 | | Legionella pneumophila SidJ/ Calmodulin 2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, Calmodulin-2, ... | | Authors: | Mao, Y, Sulpizio, A, Minelli, M.E, Wu, X. | | Deposit date: | 2019-07-01 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ.
Elife, 8, 2019
|
|
6CMY
 
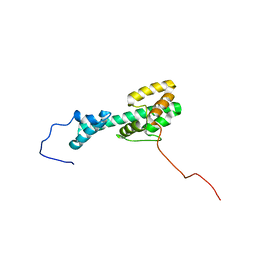 | | Solution NMR Structure Determination of Mouse Melanoregulin | | Descriptor: | Melanoregulin | | Authors: | Rout, A.K, Wu, X, Strub, M.P, Starich, M.R, Hammer III, J.A, Tjandra, N. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Melanoregulin Reveals a Role for Cholesterol Recognition in the Protein's Ability to Promote Dynein Function.
Structure, 26, 2018
|
|
8EFR
 
 | | CryoEM of the soluble OPA1 interfaces with GDP-AlFx bound from the helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.48 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
2BYD
 
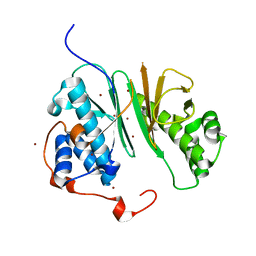 | | Structure of aminoadipate-semialdehyde dehydrogenase- phosphopantetheinyl transferase | | Descriptor: | BROMIDE ION, HSPC223 | | Authors: | Bunkoczi, G, Wu, X, Dubinina, E, Johansson, C, Smee, C, Turnbull, A, Oppermann, U, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J. | | Deposit date: | 2005-07-29 | | Release date: | 2005-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and substrate recognition of human holo ACP synthase.
Chem. Biol., 14, 2007
|
|
2C43
 
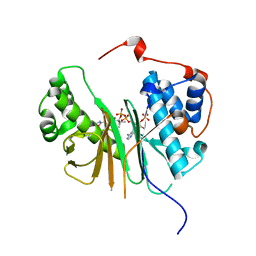 | | STRUCTURE OF AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE- PHOSPHOPANTETHEINYL TRANSFERASE IN COMPLEX WITH COENZYME A | | Descriptor: | AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE-PHOSPHOPANTETHEINYL TRANSFERASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Bunkoczi, G, Wu, X, Dubinina, E, Johansson, C, Smee, C, Turnbull, A, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Oppermann, U. | | Deposit date: | 2005-10-14 | | Release date: | 2005-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism and substrate recognition of human holo ACP synthase.
Chem. Biol., 14, 2007
|
|
2BEL
 
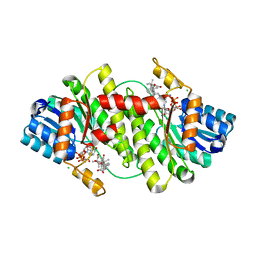 | | Structure of human 11-beta-hydroxysteroid dehydrogenase in complex with NADP and carbenoxolone | | Descriptor: | CARBENOXOLONE, CHLORIDE ION, CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1, ... | | Authors: | Kavanagh, K, Wu, X, Svensson, S, Elleby, B, von Delft, F, Debreczeni, J.E, Sharma, S, Bray, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Abrahmsen, L, Oppermann, U. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The High Resolution Structures of Human, Murine and Guinea Pig 11-Beta-Hydroxysteroid Dehydrogenase Type 1 Reveal Critical Differences in Active Site Architecture
To be Published
|
|
7X8R
 
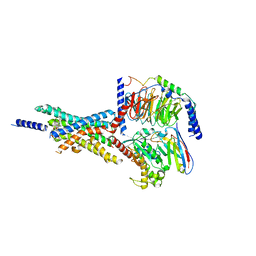 | | Cryo-EM structure of the Boc5-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-[(2-methylpropan-2-yl)oxycarbonylamino]phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X8S
 
 | | Cryo-EM structure of the WB4-24-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-(2-methylpropanoylamino)phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6U07
 
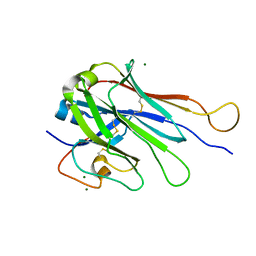 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
7JWL
 
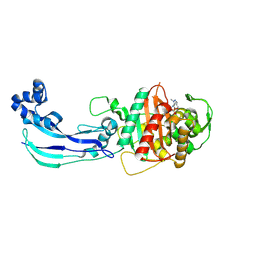 | | Crystal Structure of Pseudomonas aeruginosa Penicillin Binding Protein 3 (PAE-PBP3) bound to ETX0462 | | Descriptor: | CHLORIDE ION, ETX0462 (Bound form), Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Mayclin, S.J, Abendroth, J, Horanyi, P.S, Sylvester, M, Wu, X, Shapiro, A, Moussa, S, Durand-Reville, T.F. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a new antibiotic class for drug-resistant infections.
Nature, 597, 2021
|
|
2C9H
 
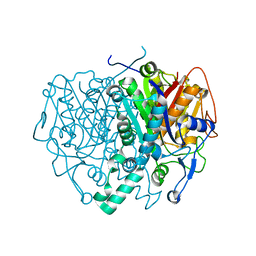 | | Structure of mitochondrial beta-ketoacyl synthase | | Descriptor: | MITOCHONDRIAL BETA-KETOACYL SYNTHASE, NICKEL (II) ION | | Authors: | Bunkoczi, G, Wu, X, Smee, C, Gileadi, O, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, von Delft, F, Oppermann, U. | | Deposit date: | 2005-12-12 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Mitochondrial Beta-Ketoacyl Synthase
To be Published
|
|
6W5S
 
 | | NPC1 structure in GDN micelles at pH 8.0 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
