5Z0Q
 
 | | Crystal Structure of OvoB | | Descriptor: | Aminotransferase, class I and II, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cai, Y.J, Huang, P, Wu, L, Zhou, J.H, Liu, P.H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | In Vitro Reconstitution of the Remaining Steps in Ovothiol A Biosynthesis: C-S Lyase and Methyltransferase Reactions.
Org. Lett., 20, 2018
|
|
6LCV
 
 | |
6IBT
 
 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclophellitol aziridine ME737 | | Descriptor: | (1~{S},2~{S},3~{S},4~{S},5~{R},6~{S})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
6IBK
 
 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclosulfamidate ME763 | | Descriptor: | (3~{a}~{R},4~{S},5~{S},6~{S},7~{R},7~{a}~{S})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydro-3~{H}-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
6IBR
 
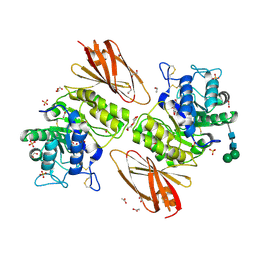 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclophellitol epoxide LWA481 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-5-(hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
6IBM
 
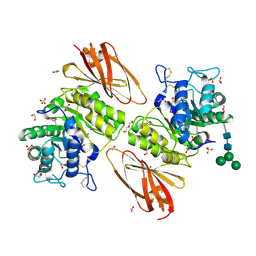 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclosulfate ME776 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
7DL0
 
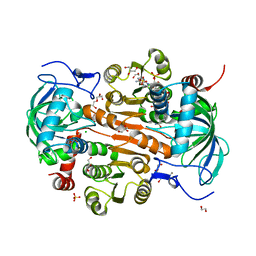 | | The mutant E310G/A314Y of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL7
 
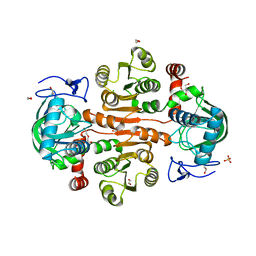 | | The wild-type structure of 3,5-DAHDHcca | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,5-diaminohexanoate dehydrogenase, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.30065823 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL1
 
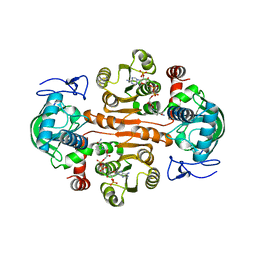 | | The mutant E310G/G323S structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL3
 
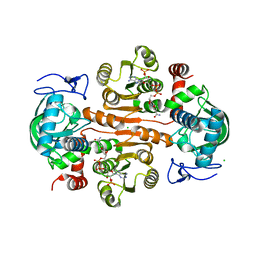 | | The structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84606934 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5YM0
 
 | | The crystal structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, SULFATE ION | | Authors: | Zang, X, Huang, W.X, Cheng, R, Wu, L, Zhou, J.H, Tang, Y, Yan, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The crystal structure of DHAD
To Be Published
|
|
5YGE
 
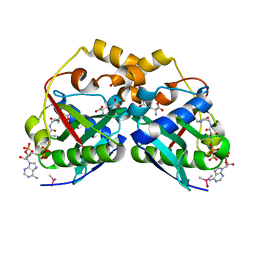 | | ArgA complexed with AceCoA and glutamate | | Descriptor: | ACETYL COENZYME *A, Amino-acid acetyltransferase, CACODYLIC ACID, ... | | Authors: | Yang, X, Wu, L, Ran, Y, Xu, A, Zhang, B, Yang, X, Zhang, R, Rao, Z, Li, J. | | Deposit date: | 2017-09-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Crystal structure of l-glutamate N-acetyltransferase ArgA from Mycobacterium tuberculosis
Biochim. Biophys. Acta, 1865, 2017
|
|
5YQT
 
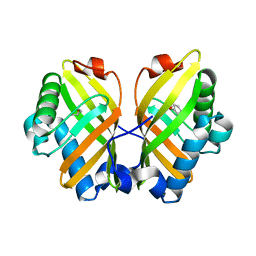 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z.T, Wu, L, Reetz, M.T, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-11-07 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations.
J. Am. Chem. Soc., 140, 2018
|
|
6JIZ
 
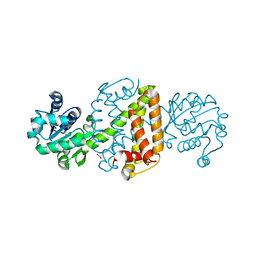 | | Apo structure of an imine reductase at 1.76 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-ethylheptane, 6-phosphogluconate dehydrogenase NAD-binding protein, ... | | Authors: | Li, H, Wu, L, Zheng, G.W, Zhou, J.H. | | Deposit date: | 2019-02-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Apo structure of an imine reductase at 1.76 Angstrom resolution
To Be Published
|
|
5Z2X
 
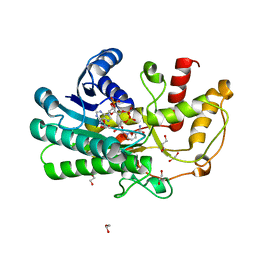 | | Structure of Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | Descriptor: | 1,2-ETHANEDIOL, Alcohol dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5X5V
 
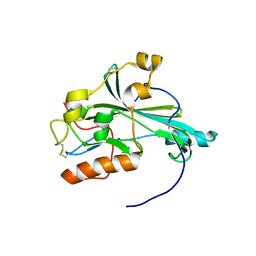 | | Crystal structure of pseudorabies virus glycoprotein D | | Descriptor: | GD | | Authors: | Li, A, Lu, G, Qi, J, Wu, L, Tian, K, Luo, T, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2017-02-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of pseudorabies virus glycoprotein D
To Be Published
|
|
5ZEC
 
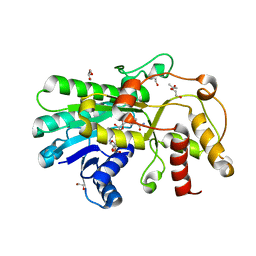 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Q136N/F161V/S196G/E214G/S237C) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wang, Y, ZHou, J.Y, Hou, X.D, Xu, G.C, Rao, Y.J, Wu, L, Zhou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5ZED
 
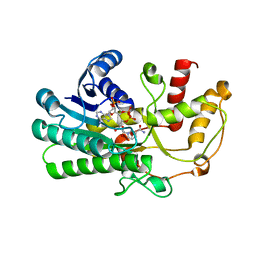 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (E214V/T215S) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein ADH | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5X5W
 
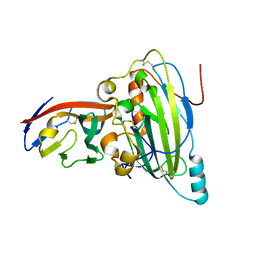 | | Crystal structure of pseudorabies virus glycoprotein D | | Descriptor: | GD, Nectin-1 | | Authors: | Li, A, Lu, G, Qi, J, Wu, L, Tian, K, Luo, T, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2017-02-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pseudorabies virus glycoprotein D
To Be Published
|
|
6B1T
 
 | | Improved cryoEM structure of human adenovirus type 5 with atomic details of minor proteins VI and VII | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Dai, X.H, Wu, L, Sun, R, Zhou, Z.H. | | Deposit date: | 2017-09-18 | | Release date: | 2017-09-27 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomic Structures of Minor Proteins VI and VII in Human Adenovirus.
J. Virol., 91, 2017
|
|
6JIT
 
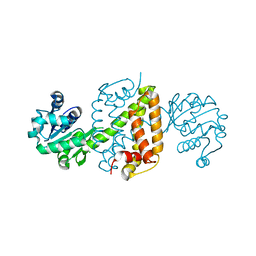 | | Complex structure of an imine reductase at 2.05 Angstrom resolution | | Descriptor: | 1-(2-phenylethyl)-3,4-dihydroisoquinoline, 6-phosphogluconate dehydrogenase NAD-binding protein, CHLORIDE ION, ... | | Authors: | Li, H, Wu, L, Zheng, G.W, Zhou, J.H. | | Deposit date: | 2019-02-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Complex structure of an imine reductase at 2.05 Angstrom resolution
To Be Published
|
|
7VYU
 
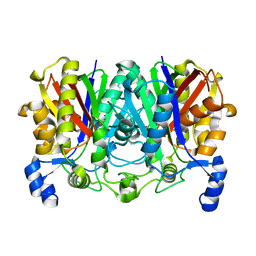 | |
6IX4
 
 | | Structure of an epoxide hydrolase from Aspergillus usamii E001 (AuEH2) at 1.51 Angstroms resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hu, D, Hu, B.C, Hou, X.D, Wu, L, Rao, Y.J, Wu, M.C. | | Deposit date: | 2018-12-09 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Nearly perfect kinetic resolution of racemic o-nitrostyrene oxide by AuEH2, a microsomal epoxide hydrolase from Aspergillus usamii, with high enantio- and regio-selectivity.
Int.J.Biol.Macromol., 169, 2021
|
|
7ETK
 
 | |
7ETL
 
 | | The crystal structure of FtmOx1-Y68F | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-05-13 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.992128 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of the Endoperoxide Synthase FtmOx1.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
