7FIY
 
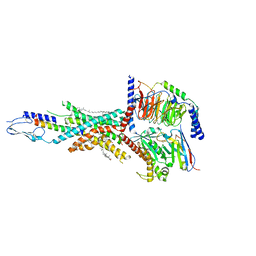 | | Cryo-EM structure of the tirzepatide-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-01 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7FIM
 
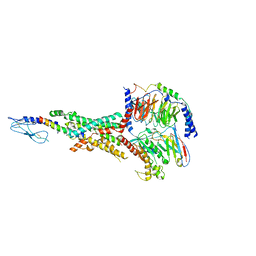 | | Cryo-EM structure of the tirzepatide (LY3298176)-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,human glucagon like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-07-31 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
2OHW
 
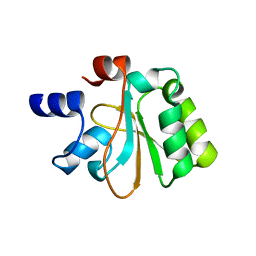 | | Crystal structure of the YueI protein from Bacillus subtilis | | Descriptor: | YueI protein | | Authors: | Bonanno, J.B, Jin, X, Mu, H, Dickey, M, Bain, K.T, Wu, B, Chen, T, Reyes, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-10 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the YueI protein from Bacillus subtilis
To be Published
|
|
2O3A
 
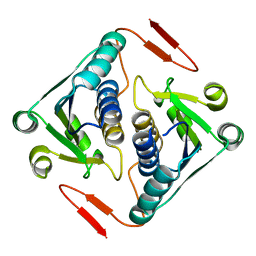 | | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus | | Descriptor: | UPF0106 protein AF_0751 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Wu, B, Slocombe, A, Sridhar, V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus
To be Published
|
|
7VJQ
 
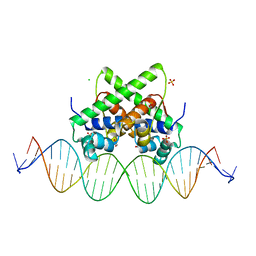 | | Pectobacterium phage ZF40 apo-aca2 complexed with 26bp DNA substrate | | Descriptor: | CHLORIDE ION, DNA (27-MER), GLYCEROL, ... | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
5Y6R
 
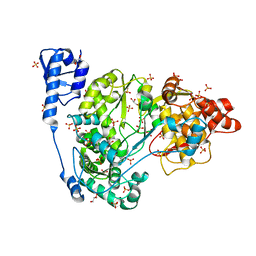 | | Crystal structure of CSFV NS5B | | Descriptor: | GLYCEROL, Genome polyprotein, SULFATE ION | | Authors: | Li, W, Wu, B. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of Classical Swine Fever Virus NS5B Reveals a Novel N-Terminal Domain
J. Virol., 92, 2018
|
|
6JNM
 
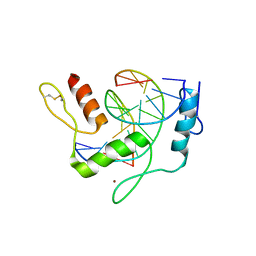 | | REF6 ZnF2-4-NAC004-mC3 complex | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*(5CM)P*TP*GP*TP*TP*TP*TP*G)-3'), Lysine-specific demethylase REF6, ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
6JNN
 
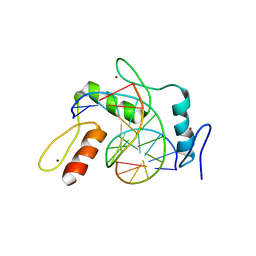 | | REF6 ZnF2-4-NAC004-mC1 complex | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*(5CM)P*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), Lysine-specific demethylase REF6, ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
7E9G
 
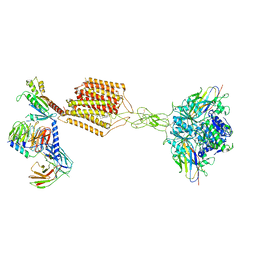 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu2 | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, DN13, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
7EPC
 
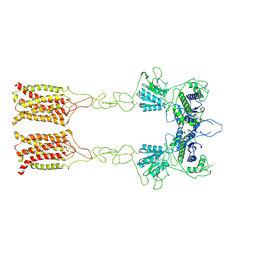 | | Cryo-EM structure of inactive mGlu7 homodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPE
 
 | | Crystal structure of mGlu2 bound to NAM563 | | Descriptor: | 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPF
 
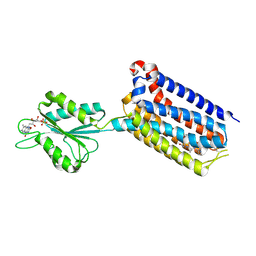 | | Crystal structure of mGlu2 bound to NAM597 | | Descriptor: | (8~{R})-4-[2,4-bis(fluoranyl)phenyl]-8-methyl-7-[(2-methylpyrazol-3-yl)methyl]-6,8-dihydro-5~{H}-1,7-naphthyridine-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7F1R
 
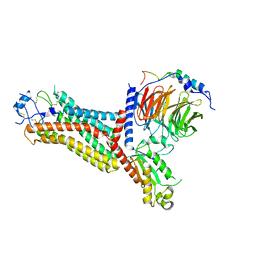 | | Cryo-EM structure of the chemokine receptor CCR5 in complex with RANTES and Gi | | Descriptor: | C-C motif chemokine 5,C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1T
 
 | | Crystal structure of the human chemokine receptor CCR5 in complex with MIP-1a | | Descriptor: | C-C motif chemokine 3,C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ZINC ION | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1Q
 
 | | Cryo-EM structure of the chemokine receptor CCR5 in complex with MIP-1a and Gi | | Descriptor: | C-C motif chemokine 3,C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7VJP
 
 | | Selenomethionine-derived Pectobacterium phage ZF40 apo-Aca2 | | Descriptor: | SULFATE ION, anti-CRISPR-associated protein Aca2 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
5YAD
 
 | | Crystal structure of Marf1 Lotus domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1, SULFATE ION | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YAA
 
 | | Crystal structure of Marf1 NYN domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1 | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A6J
 
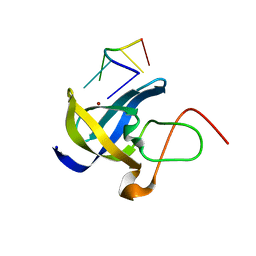 | | Crystal structure of Zebra fish Y-box protein1 (YB-1) Cold-shock domain in complex with 6mer m5C RNA | | Descriptor: | RNA (5'-R(P*CP*AP*UP*(5MC)P*U)-3'), ZINC ION, Zebra fish Y-box protein1 (YB-1) | | Authors: | Zhang, M.M, Wu, B.X, Huang, Y, Ma, J.B. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Mol.Cell, 75, 2019
|
|
7EPA
 
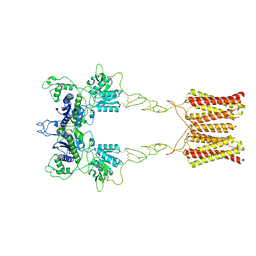 | | Cryo-EM structure of inactive mGlu2 homodimer | | Descriptor: | Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPD
 
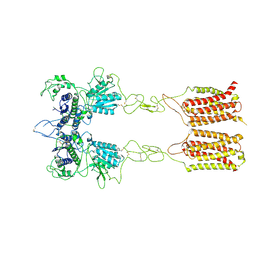 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
2OZ3
 
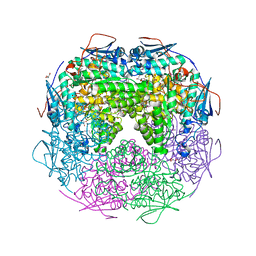 | | Crystal structure of L-Rhamnonate dehydratase from Azotobacter vinelandii | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wu, B, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Rhamnonate dehydratase from azotobacter vinelandii
To be Published
|
|
6AKX
 
 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 21 | | Descriptor: | C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-2-yl)propyl]cyclopentanecarboxamide, NITRATE ION, ... | | Authors: | Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
2OZ8
 
 | | Crystal structure of putative mandelate racemase from Mesorhizobium loti | | Descriptor: | Mll7089 protein, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-25 | | Release date: | 2007-03-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of putative mandelate racemase from Mesorhizobium loti
To be Published
|
|
7EPB
 
 | | Cryo-EM structure of LY354740-bound mGlu2 homodimer | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, Anti-RON nanobody, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
