2Q71
 
 | | Uroporphyrinogen Decarboxylase G168R single mutant enzyme in complex with coproporphyrinogen-III | | Descriptor: | COPROPORPHYRINOGEN III, Uroporphyrinogen decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Stadtmueller, B.M, Edwards, C.Q, Hill, C.P, Kushner, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two Novel Uropophyrinogen Decarboxylase (URO-D) Mutations Causing Hepatoerythropoietic Porphyria (HEP)
Transl.Res., 149, 2007
|
|
4LCB
 
 | | Structure of Vps4 homolog from Acidianus hospitalis | | Descriptor: | CHLORIDE ION, Cell division protein CdvC, Vps4 | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-21 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4LGM
 
 | | Crystal Structure of Sulfolobus solfataricus Vps4 | | Descriptor: | CHLORIDE ION, Vps4 AAA ATPase | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
1QC7
 
 | | T. MARITIMA FLIG C-TERMINAL DOMAIN | | Descriptor: | PROTEIN (FLIG) | | Authors: | Lloyd, S.A, Whitby, F.G, Blair, D, Hill, C.P. | | Deposit date: | 1999-05-18 | | Release date: | 1999-08-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor
Nature, 400, 1999
|
|
1R3Y
 
 | | Uroporphyrinogen Decarboxylase in complex with coproporphyrinogen-III | | Descriptor: | COPROPORPHYRINOGEN III, Uroporphyrinogen Decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Kushner, J.P, Hill, C.P. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase
Embo J., 22, 2003
|
|
1R3V
 
 | | Uroporphyrinogen Decarboxylase single mutant D86E in complex with coproporphyrinogen-I | | Descriptor: | BETA-MERCAPTOETHANOL, COPROPORPHYRINOGEN I, Uroporphyrinogen Decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Kushner, J.P, Hill, C.P. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase
Embo J., 22, 2003
|
|
7SMU
 
 | | Crystal Structure of Consomatin-Ro1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45-pentadecaoxaoctatetracontane-1,48-diol, Consomatin-Ro1 | | Authors: | Ramiro, I.B.L, Whitby, F.G, Hill, C.P, Safavi-Hemami, H, Concepcion, G.P, Olivera, B.M. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads.
Sci Adv, 8, 2022
|
|
7TXE
 
 | | Plasmodium falciparum Cyt c2 DSD | | Descriptor: | Cytochrome c2, HEME C | | Authors: | Hill, C.P, Wienkers, H.J, Whitby, F.G. | | Deposit date: | 2022-02-08 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct tests of cytochrome c and c1 functions in the electron transport chain of malaria parasites
Proc Natl Acad Sci U S A, 120, 2023
|
|
7U2V
 
 | |
5KKE
 
 | |
5KLU
 
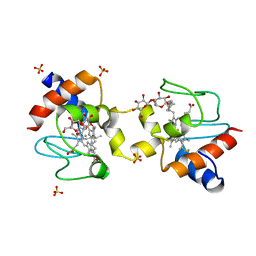 | |
3B6H
 
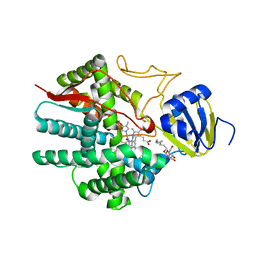 | | Crystal structure of human prostacyclin synthase in complex with inhibitor minoxidil | | Descriptor: | 6-PIPERIDIN-1-YLPYRIMIDINE-2,4-DIAMINE 3-OXIDE, PROTOPORPHYRIN IX CONTAINING FE, Prostacyclin synthase, ... | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
3B99
 
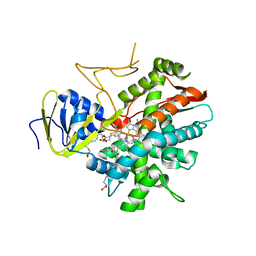 | | Crystal structure of zebrafish prostacyclin synthase (cytochrome P450 8A1) in complex with substrate analog U51605 | | Descriptor: | (5Z)-7-{(1R,4S,5R,6R)-6-[(1E)-oct-1-en-1-yl]-2,3-diazabicyclo[2.2.1]hept-2-en-5-yl}hept-5-enoic acid, PROTOPORPHYRIN IX CONTAINING FE, Prostaglandin I2 synthase | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-11-03 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
3B98
 
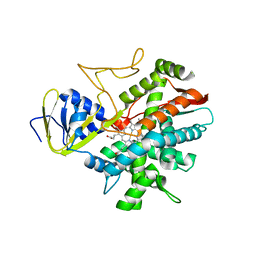 | | Crystal structure of zebrafish prostacyclin synthase (cytochrome P450 8A1) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Prostaglandin I2 synthase | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-11-03 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
1JPI
 
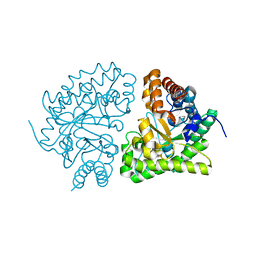 | | Phe232Leu mutant of human UROD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
1JPK
 
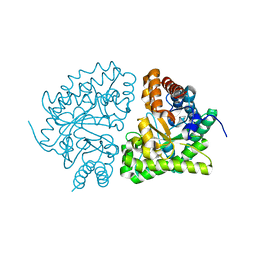 | | Gly156Asp mutant of Human UroD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
1JPH
 
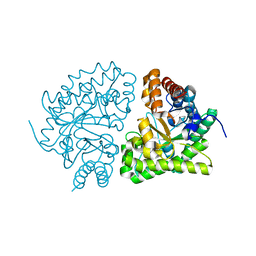 | | Ile260Thr mutant of Human UroD, human uroporphyrinogen III decarboxylase | | Descriptor: | UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Phillips, J.D, Parker, T.L, Schubert, H.L, Whitby, F.G, Hill, C.P, Kushner, J.P. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional consequences of naturally occurring mutations in human uroporphyrinogen decarboxylase.
Blood, 98, 2001
|
|
4V7O
 
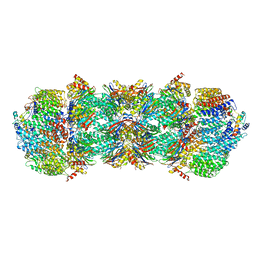 | | Proteasome Activator Complex | | Descriptor: | Proteasome activator BLM10, Proteasome component C1, Proteasome component C11, ... | | Authors: | Hill, C.P, Whitby, F.G. | | Deposit date: | 2009-12-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening.
Mol.Cell, 37, 2010
|
|
3L37
 
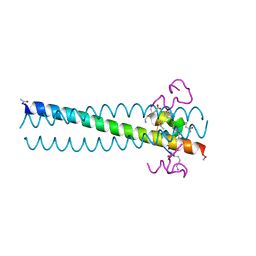 | | PIE12 D-peptide against HIV entry | | Descriptor: | GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
3L35
 
 | | PIE12 D-peptide against HIV entry | | Descriptor: | GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
3L36
 
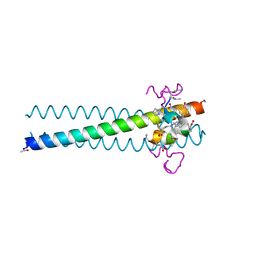 | | PIE12 D-peptide against HIV entry | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
4WLR
 
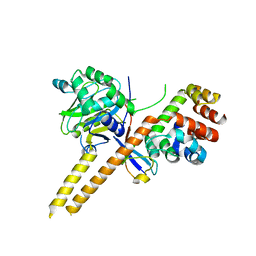 | | Crystal Structure of mUCH37-hRPN13 CTD-hUb complex | | Descriptor: | Polyubiquitin-B, Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
4WLP
 
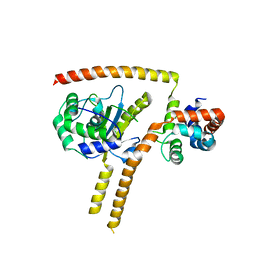 | | Crystal structure of UCH37-NFRKB Inhibited Deubiquitylating Complex | | Descriptor: | Nuclear factor related to kappa-B-binding protein, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
4WNN
 
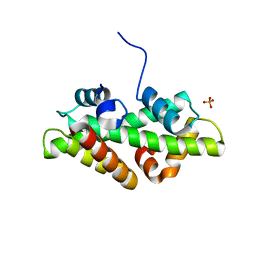 | | SPT16-H2A-H2B FACT HISTONE Complex | | Descriptor: | Histone H2A.1, Histone H2B.1, PHOSPHATE ION, ... | | Authors: | Kemble, D.J, Hill, C.P, Whitby, F.G, Formosa, T, McCullough, L.L. | | Deposit date: | 2014-10-13 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs.
Mol.Cell, 60, 2015
|
|
4WLQ
 
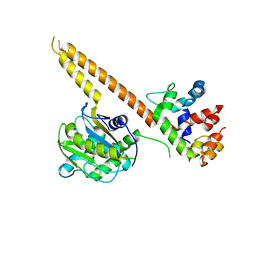 | | Crystal structure of mUCH37-hRPN13 CTD complex | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
