1W1I
 
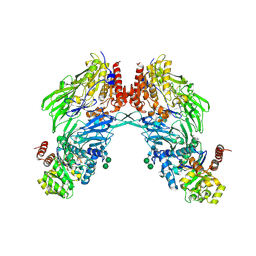 | | Crystal structure of dipeptidyl peptidase IV (DPPIV or CD26) in complex with adenosine deaminase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2004-06-22 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface.
J. Biol. Chem., 279, 2004
|
|
6M8Q
 
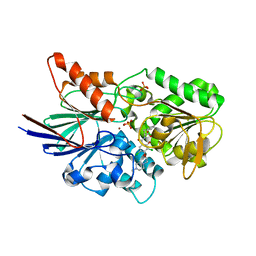 | | Cleavage and Polyadenylation Specificity Factor Subunit 3 (CPSF3) in complex with NVP-LTM531 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 3, N-{3,5-dichloro-2-hydroxy-4-[2-(4-methylpiperazin-1-yl)ethoxy]benzene-1-carbonyl}-L-phenylalanine, PHOSPHATE ION, ... | | Authors: | Weihofen, W.A, Salcius, M, Michaud, G. | | Deposit date: | 2018-08-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing's sarcoma.
Nat.Chem.Biol., 16, 2020
|
|
6PEB
 
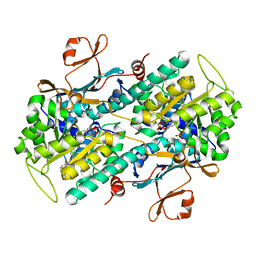 | | Crystal Structure of human NAMPT in complex with NVP-LTM976 | | Descriptor: | N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-3-(pyridin-3-yl)azetidine-1-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Weihofen, W.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Scaffold Morphing Identifies 3-Pyridyl Azetidine Ureas as Inhibitors of Nicotinamide Phosphoribosyltransferase (NAMPT).
Acs Med.Chem.Lett., 10, 2019
|
|
6USY
 
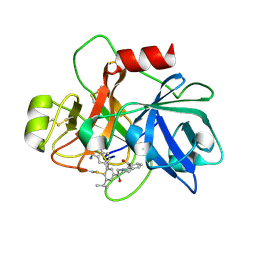 | | COAGULATION FACTOR XI CATALYTIC DOMAIN (C123S) IN COMPLEX WITH NVP-XIV936 | | Descriptor: | 1-[(2S)-2-{3-[(3S)-3-amino-2,3-dihydro-1-benzofuran-5-yl]-5-(propan-2-yl)phenyl}-2-hydroxyethyl]-1H-indole-7-carboxylic acid, Coagulation factor XIa light chain | | Authors: | Weihofen, W.A, Clark, K, Nunes, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6B76
 
 | | Crystal Structure of human NAMPT in complex with NVP-LVR596 | | Descriptor: | (1S,2S)-N-{4-[(1S)-1-(propanoylamino)ethyl]phenyl}-2-(pyridin-3-yl)cyclopropane-1-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
6B75
 
 | | Crystal Structure of human NAMPT in complex with NVP-LOQ594 | | Descriptor: | 4-[(piperazin-1-yl)methyl]-N-{[4-({[(pyridin-3-yl)methyl]carbamoyl}amino)phenyl]methyl}benzamide, Nicotinamide phosphoribosyltransferase | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
6ATB
 
 | | Crystal Structure of human NAMPT in complex with NVP-LOD812 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-N'-[(pyridin-3-yl)methyl]urea, ... | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-08-28 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
2BGN
 
 | | HIV-1 Tat protein derived N-terminal nonapeptide Trp2-Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-03 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
2BGR
 
 | | Crystal structure of HIV-1 Tat derived nonapeptides Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-04 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
6AZJ
 
 | | Crystal Structure of human NAMPT in complex with NVP-LQN520 | | Descriptor: | (1S,2S)-N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-2-(pyridin-3-yl)cyclopropane-1-carboxamide, Nicotinamide phosphoribosyltransferase | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-09-11 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
2CH5
 
 | | Crystal structure of human N-acetylglucosamine kinase in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Weihofen, W.A, Berger, M, Chen, H, Saenger, W, Hinderlich, S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-09-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with Adp/Glucose: Insights Into Substrate Specificity and Regulation.
J.Mol.Biol., 364, 2006
|
|
2CH6
 
 | | Crystal structure of human N-acetylglucosamine kinase in complex with ADP and glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N-ACETYL-D-GLUCOSAMINE KINASE, alpha-D-glucopyranose | | Authors: | Weihofen, W.A, Berger, M, Chen, H, Saenger, W, Hinderlich, S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structures of Human N-Acetylglucosamine Kinase in Two Complexes with N-Acetylglucosamine and with Adp/Glucose: Insights Into Substrate Specificity and Regulation.
J.Mol.Biol., 364, 2006
|
|
2CAX
 
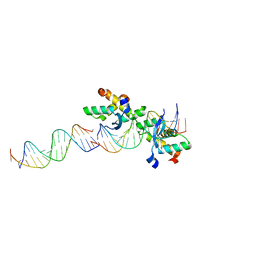 | | STRUCTURAL BASIS FOR COOPERATIVE BINDING OF RIBBON-HELIX-HELIX REPRESSOR OMEGA TO MUTATED DIRECT DNA HEPTAD REPEATS | | Descriptor: | 5'-D(*CP*TP*TP*GP*TP*GP*AP*CP*TP*TP *GP*TP*GP*AP*TP*TP*CP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*AP *TP*CP*AP*CP*AP*AP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*GP *TP*CP*AP*CP*AP*AP*GP*C)-3', ... | | Authors: | Weihofen, W.A, Cicek, A, Pratto, F, Alonso, J.C, Saenger, W. | | Deposit date: | 2005-12-23 | | Release date: | 2006-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Nucleic Acids Res., 34, 2006
|
|
2BNZ
 
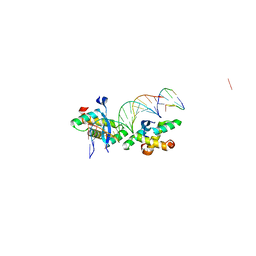 | | Structural basis for cooperative binding of Ribbon-Helix-Helix Omega repressor to inverted DNA heptad repeats | | Descriptor: | 5'-D(*CP*TP*AP*AP*TP*CP*AP*CP*TP*TP *GP*TP*GP*AP*TP*TP*CP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*GP *TP*GP*AP*TP*TP*AP*GP*C)-3', ORF OMEGA | | Authors: | Weihofen, W.A, Cicek, A, Pratto, F, Alonso, J.C, Saenger, W. | | Deposit date: | 2005-04-06 | | Release date: | 2006-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Nucleic Acids Res., 34, 2006
|
|
2BNW
 
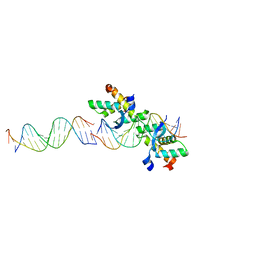 | | Structural basis for cooperative binding of Ribbon-Helix-Helix Omega repressor to direct DNA heptad repeats | | Descriptor: | 5'-D(*CP*TP*TP*GP*TP*GP*AP*TP*TP*TP *GP*TP*GP*AP*TP*TP*CP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*AP *TP*CP*AP*CP*AP*AP*GP*C)-3', ORF OMEGA | | Authors: | Weihofen, W.A, Cicek, A, Pratto, F, Alonso, J.C, Saenger, W. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Nucleic Acids Res., 34, 2006
|
|
2WDT
 
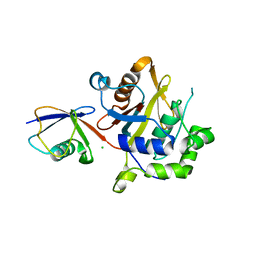 | | Crystal structure of Plasmodium falciparum UCHL3 in complex with the suicide inhibitor UbVME | | Descriptor: | CHLORIDE ION, UBIQUITIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE L3 | | Authors: | Weihofen, W.A, Artavanis-Tsakonas, K, Gaudet, R, Ploegh, H.L. | | Deposit date: | 2009-03-26 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and Structural Studies of the Plasmodium Falciparum Ubiquitin and Nedd8 Hydrolase Uchl3.
J.Biol.Chem., 285, 2010
|
|
2WE6
 
 | |
8VQC
 
 | | Structure of the voltage-gated sodium channel NavPas from American Cockroach Periplaneta Americana in complex with scorpion alpha-toxin LqhaIT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-insect toxin LqhaIT, ... | | Authors: | Phulera, S, Khoshouei, M, Whicher, J, Weihofen, W.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Scorpion alpha-toxin Lqh alpha IT specifically interacts with a glycan at the pore domain of voltage-gated sodium channels.
Structure, 32, 2024
|
|
2OZE
 
 | | The Crystal structure of Delta protein of pSM19035 from Streptoccocus pyogenes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Orf delta', ... | | Authors: | Cicek, A, Weihofen, W, Saenger, W. | | Deposit date: | 2007-02-26 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation.
Nucleic Acids Res., 36, 2008
|
|
1GSZ
 
 | | Crystal Structure of a Squalene Cyclase in Complex with the Potential Anticholesteremic Drug Ro48-8071 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE, [4-({6-[ALLYL(METHYL)AMINO]HEXYL}OXY)-2-FLUOROPHENYL](4-BROMOPHENYL)METHANONE | | Authors: | Lenhart, A, Weihofen, W.A, Pleschke, A.E.W, Schulz, G.E. | | Deposit date: | 2002-01-09 | | Release date: | 2003-01-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Squalene Cyclase in Complex with the Potential Anticholesteremic Drug Ro48-8071
Chem.Biol., 9, 2002
|
|
5KTE
 
 | | Crystal structure of Deinococcus radiodurans MntH, an Nramp-family transition metal transporter | | Descriptor: | Divalent metal cation transporter MntH, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Bane, L.B, Gaudet, R, Weihofen, W.A, Singharoy, A. | | Deposit date: | 2016-07-11 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.941 Å) | | Cite: | Crystal Structure and Conformational Change Mechanism of a Bacterial Nramp-Family Divalent Metal Transporter.
Structure, 24, 2016
|
|
2J7Q
 
 | | Crystal structure of the ubiquitin-specific protease encoded by murine cytomegalovirus tegument protein M48 in complex with a ubquitin-based suicide substrate | | Descriptor: | GLYCEROL, MAGNESIUM ION, MCMV TEGUMENT PROTEIN M48 ENCODED UBIQUITIN- SPECIFIC PROTEASE, ... | | Authors: | Schlieker, C, Weihofen, W.A, Frijns, E, Kattenhorn, L.M, Gaudet, R, Ploegh, H.L. | | Deposit date: | 2006-10-16 | | Release date: | 2007-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Herpesvirus-Encoded Cysteine Protease Reveals a Unique Class of Deubiquitinating Enzymes
Mol.Cell, 25, 2007
|
|
4ZI8
 
 | | Structure of mouse clustered PcdhgC3 EC1-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Nicoludis, J.M, Lau, S.-Y, Scharfe, C.P.I, Marks, D.S, Weihofen, W.A, Gaudet, R. | | Deposit date: | 2015-04-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity.
Structure, 23, 2015
|
|
4ZI9
 
 | | Structure of mouse clustered PcdhgA1 EC1-3 | | Descriptor: | CALCIUM ION, MCG133388, isoform CRA_t | | Authors: | Nicoludis, J.M, Lau, S.-Y, Scharfe, C.P.I, Marks, D.S, Weihofen, W.A, Gaudet, R. | | Deposit date: | 2015-04-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity.
Structure, 23, 2015
|
|
1H3A
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (2E)-N-ALLYL-4-{[3-(4-BROMOPHENYL)-5-FLUORO-1-METHYL-1H-INDAZOL-6-YL]OXY}-N-METHYL-2-BUTEN-1-AMINE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
