6VFJ
 
 | |
6VFH
 
 | |
6VFK
 
 | |
6V0R
 
 | | BG505 SOSIP.664 Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nogal, B, Cottrell, C.A, Ward, A.B. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Mapping Polyclonal Antibody Responses in Non-human Primates Vaccinated with HIV Env Trimer Subunit Vaccines.
Cell Rep, 30, 2020
|
|
6VFI
 
 | |
6VFL
 
 | |
8SW4
 
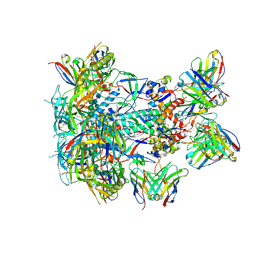 | | BG505 GT1.1 SOSIP in complex with NHP Fabs 21N13, 21M20 and RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 21M20 heavy chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Zhang, S, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Germline-targeting HIV vaccination induces neutralizing antibodies to the CD4 binding site.
Sci Immunol, 9, 2024
|
|
8SW3
 
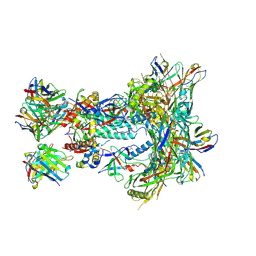 | | BG505 GT1.1 SOSIP in complex with NHP Fabs 12C11 and RM20A3 | | Descriptor: | 12C11 heavy chain variable region, 12C11 light chain variable region, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Torres, J.L, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Germline-targeting HIV vaccination induces neutralizing antibodies to the CD4 binding site.
Sci Immunol, 9, 2024
|
|
8TP6
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting Fab 4-1-1E02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 4-1-1E02 Fab, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP7
 
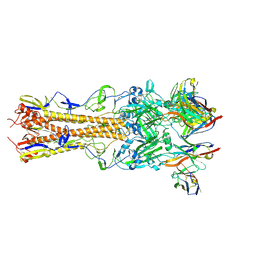 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with Sa-targeting Fab 4-1-1G03 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 4-1-1G03, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP9
 
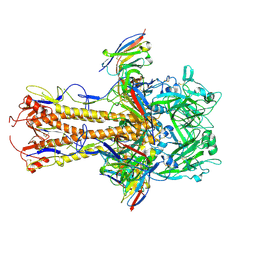 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with medial-junction-targeting Fab 2-2-1G06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 2-2-1G06, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP3
 
 | | H1 hemagglutinin (NC99) in complex with RBS-targeting Fab 1-1-1F05 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1F05, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TPA
 
 | | H1 hemagglutinin (NC99) in complex with medial-junction-targeting Fab 2-2-1G06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 2-2-1G06, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP4
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting Fab 1-1-1E04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1E04, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP2
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting 1-1-1F05 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1F05, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP5
 
 | | H1 hemagglutinin (NC99) in complex with RBS-targeting Fab 1-1-1E04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1E04, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
7JJI
 
 | | Structure of SARS-CoV-2 3Q-2P full-length prefusion spike trimer (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
7JJJ
 
 | | Structure of SARS-CoV-2 3Q-2P full-length dimers of spike trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bangaru, S, Turner, H.L, Ozorowski, G, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2020-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.
Science, 370, 2020
|
|
8T4D
 
 | | MD65 N332-GT5 SOSIP in complex with RM_N332_08 Fab and RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MD65 N332-GT5 SOSIP gp120, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates.
Science, 384, 2024
|
|
8T49
 
 | | MD65 N332-GT5 SOSIP in complex with RM_N332_03 Fab and RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MD65 N332-GT5 SOSIP gp120, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates.
Science, 384, 2024
|
|
8SWW
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody IF3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NHP polyclonal antibody IF3 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8SX3
 
 | | 10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10 | | Descriptor: | 10E8 Fab heavy chain, 10E8 light chain, 10E8-GT10.2 immunogen, ... | | Authors: | Huang, J, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Vaccination induces broadly neutralizing antibody precursors to HIV gp41.
Nat.Immunol., 25, 2024
|
|
8T4B
 
 | | MD65 N332-GT5 SOSIP in complex with RM_N332_32 Fab and RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MD65 N332-GT5 SOSIP gp120, MD65 N332-GT5 SOSIP gp41, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates.
Science, 384, 2024
|
|
8T4K
 
 | | MD64 N332-GT5 SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MD64 N332-GT5 SOSIP gp120, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates.
Science, 384, 2024
|
|
8T4A
 
 | | MD65 N332-GT5 SOSIP in complex with RM_N332_36 Fab and RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MD65 N332-GT5 SOSIP gp120, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates.
Science, 384, 2024
|
|
