4ELQ
 
 | | Ferric binding protein with carbonate | | Descriptor: | CARBONATE ION, Iron ABC transporter, periplasmic iron-binding protein | | Authors: | Wang, Q, Liu, X.Q, Wang, X.Q. | | Deposit date: | 2012-04-11 | | Release date: | 2013-04-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Crystal structure of ferric binding protein A
To be Published
|
|
1YX5
 
 | |
1YX6
 
 | |
9LXN
 
 | | Human RNA Polymerase III de novo transcribing complex 5 (TC5)(without 3'dATP) | | Descriptor: | DNA (127-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2025-02-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action
Nature, 2025
|
|
9LXO
 
 | | Human RNA Polymerase III de novo transcribing complex 6 (TC6)(without 3'dATP) | | Descriptor: | DNA (127-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2025-02-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action
Nature, 2025
|
|
4Y93
 
 | | Crystal structure of the PH-TH-kinase construct of Bruton's tyrosine kinase (Btk) | | Descriptor: | 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, CALCIUM ION, Non-specific protein-tyrosine kinase,Non-specific protein-tyrosine kinase, ... | | Authors: | Wang, Q, Kuriyan, J. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
3SVN
 
 | |
3SVU
 
 | |
3SVR
 
 | |
5XYB
 
 | |
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | Descriptor: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | Deposit date: | 2017-07-24 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
1FYA
 
 | |
1FY9
 
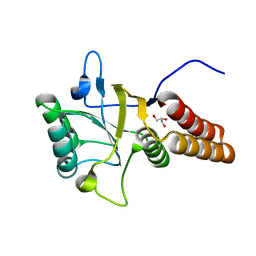 | |
8YRH
 
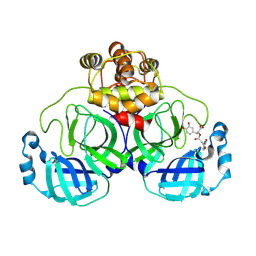 | | Complex of SARS-CoV-2 main protease and Rosmarinic acid | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, 3C-like proteinase nsp5 | | Authors: | Wang, Q.S, Li, Q.H. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Structural basis of rosmarinic acid inhibitory mechanism on SARS-CoV-2 main protease.
Biochem.Biophys.Res.Commun., 724, 2024
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
9K3V
 
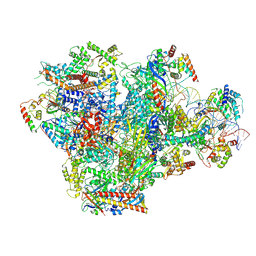 | | Human RNA Polymerase III de novo transcribing complex 4 (TC4) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (96-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action
Nature, 2025
|
|
9K36
 
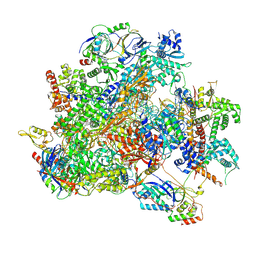 | | Human RNA Polymerase III de novo transcribing complex 12 (TC12) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (54-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action
Nature, 2025
|
|
9K39
 
 | | Human RNA Polymerase III de novo transcribing complex 8 (TC8) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (50-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action
Nature, 2025
|
|
9K2G
 
 | | Human RNA Polymerase III de novo transcribing complex 13 (TC13) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (104-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-17 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action
Nature, 2025
|
|
9K3U
 
 | | Human RNA Polymerase III de novo transcribing complex 5 (TC5) | | Descriptor: | DNA (97-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action
Nature, 2025
|
|
9K38
 
 | | Human RNA Polymerase III de novo transcribing complex 6 (TC6) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (51-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action
Nature, 2025
|
|
9K3B
 
 | | Human RNA Polymerase III de novo transcribing complex 10 overall (TC10-overall) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (102-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action
Nature, 2025
|
|
4Y95
 
 | | Crystal structure of the kinase domain of Bruton's tyrosine kinase with mutations in the activation loop | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, BETA-MERCAPTOETHANOL, ... | | Authors: | Wang, Q, Rosen, C.E, Kuriyan, J. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
4Y94
 
 | |
