8EAU
 
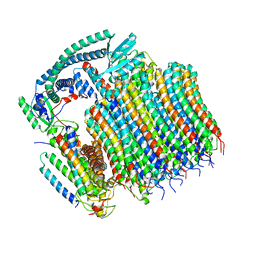 | | Yeast VO in complex with Vma21p | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform, V-type proton ATPase subunit c, ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8Y7Y
 
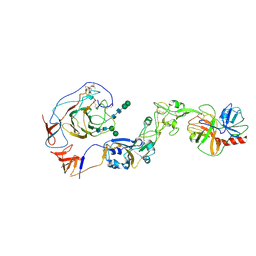 | | Local structure of HCoV-HKU1A spike in complex with TMPRSS2 and glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, H.F, Zhang, X, Lu, Y, Liu, X, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8B
 
 | | Local structure of HCoV-HKU1C spike in complex with TMPRSS2 and glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, H.F, Zhang, X, Lu, Y.C, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
5WH5
 
 | |
6LMJ
 
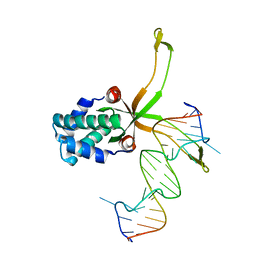 | | ASFV pA104R in complex with double-strand DNA | | Descriptor: | A104R, DNA (5'-D(*TP*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*A)-3') | | Authors: | Wang, H, Qi, J, Chai, Y, Gao, F, Liu, R. | | Deposit date: | 2019-12-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of African swine fever virus pA104R binding to DNA and its inhibition by stilbene derivatives.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6M89
 
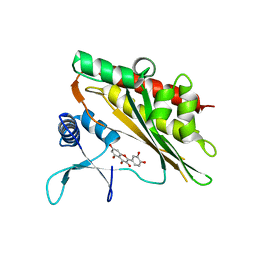 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase in complex with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2018-08-21 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inhibition of Inositol Polyphosphate Kinases by Quercetin and Related Flavonoids: A Structure-Activity Analysis.
J. Med. Chem., 62, 2019
|
|
8JJ2
 
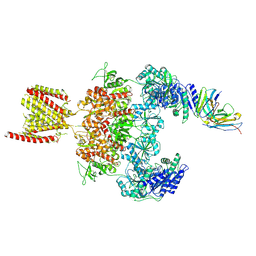 | | Cryo-EM structure of GluN1-2A NMDAR in complex with human Fab2G7 in one fab conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab2G7 Heavy Chain, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat.Struct.Mol.Biol., 2024
|
|
8JJ0
 
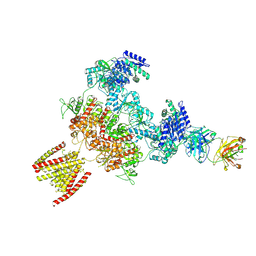 | |
8JIZ
 
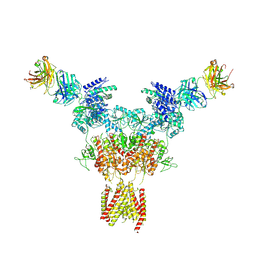 | |
8JJ1
 
 | | Cryo-EM structure of GluN1-2A NMDAR in complex with human Fab2G7 in two fab conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2G7 Heavy Chain, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat.Struct.Mol.Biol., 2024
|
|
6M8D
 
 | |
6LMH
 
 | |
6M8B
 
 | |
6M88
 
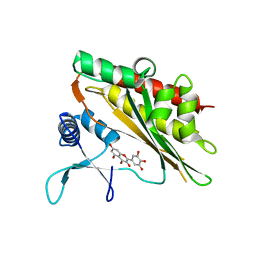 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2018-08-21 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of Inositol Polyphosphate Kinases by Quercetin and Related Flavonoids: A Structure-Activity Analysis.
J. Med. Chem., 62, 2019
|
|
8BZN
 
 | | SARS-CoV-2 non-structural protein 10 (nsp10) variant T102I | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab, ... | | Authors: | Wang, H, Rizvi, S.R.A, Dong, D, Lou, J, Wang, Q, Sopipong, W, Najar, F, Agarwal, P.K, Kozielski, F, Haider, S. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Emerging variants of SARS-CoV-2 NSP10 highlight strong functional conservation of its binding to two non-structural proteins, NSP14 and NSP16.
Elife, 12, 2023
|
|
8K3K
 
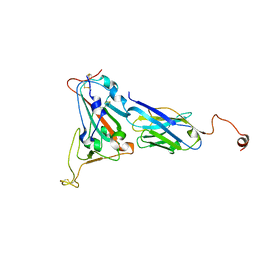 | |
4RGW
 
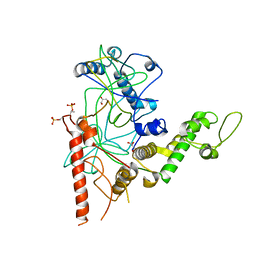 | | Crystal Structure of a TAF1-TAF7 Complex in Human Transcription Factor IID | | Descriptor: | GLYCEROL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7 | | Authors: | Wang, H, Curran, E.C, Hinds, T.R, Wang, E.H, Zheng, N. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module.
Cell Res., 24, 2014
|
|
2CSL
 
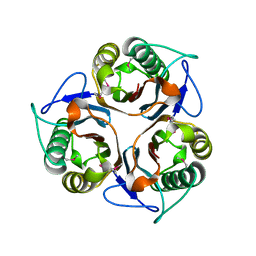 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | protein translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Jin, Z, Chrzas, J, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-22 | | Release date: | 2005-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
To be Published
|
|
6HBA
 
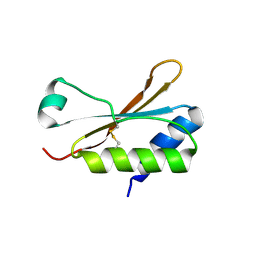 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
2DY1
 
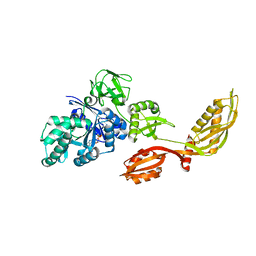 | | Crystal structure of EF-G-2 from Thermus thermophilus | | Descriptor: | Elongation factor G, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, H, Takemoto, C, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-04 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of EF-G-2 from Thermus thermophilus
To be Published
|
|
6LK5
 
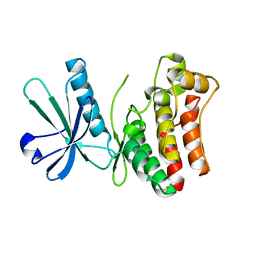 | | MLKL mutant - T357ES358D | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Wang, H.Y, Li, S, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling.
Cell Death Dis, 12, 2021
|
|
5AYR
 
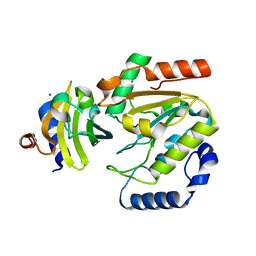 | | The crystal structure of SAUGI/human UDG complex | | Descriptor: | MAGNESIUM ION, Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
5AYS
 
 | | Crystal structure of SAUGI/HSV UDG complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
8J8Y
 
 | |
6FLH
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, SOD1 7+7, apo form | | Descriptor: | GLYCEROL, SULFATE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Yang, F, Logan, D, Oliveberg, M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Cost of Long Catalytic Loops in Folding and Stability of the ALS-Associated Protein SOD1.
J.Am.Chem.Soc., 140, 2018
|
|
