5F70
 
 | | Crystal structures of human TIM members | | Descriptor: | Hepatitis A virus cellular receptor 1 | | Authors: | Gao, G.F, Lu, G, Wang, H, Qi, J. | | Deposit date: | 2015-12-07 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of human TIM members: Ebolavirus entry-enhancing receptors
Chin.Sci.Bull., 60, 2015
|
|
5F71
 
 | | Human T-cell immunoglobulin and mucin domain protein 3 (hTIM-3) | | Descriptor: | Hepatitis A virus cellular receptor 2, SODIUM ION | | Authors: | Gao, G.F, Lu, G, Wang, H, Qi, J. | | Deposit date: | 2015-12-07 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Crystal structures of human TIM members: Ebolavirus entry-enhancing receptors
Chin.Sci.Bull., 60, 2015
|
|
4E71
 
 | | Crystal structure of the RHO GTPASE binding domain of Plexin B2 | | Descriptor: | Plexin-B2, SODIUM ION | | Authors: | Guan, X, Wang, H, Tempel, W, Tong, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the RHO GTPASE binding domain of Plexin B2
to be published
|
|
4E74
 
 | | Crystal structure of the RHO GTPASE BINDING DOMAIN of Plexin A4A | | Descriptor: | Plexin-A4, UNKNOWN ATOM OR ION | | Authors: | Guan, X, Wang, H, Tempel, W, Dong, A, Tong, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the RHO GTPASE BINDING DOMAIN of Plexin A4A
to be published
|
|
3NV5
 
 | | Crystal Structure of Cytochrome P450 CYP101D2 | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
2KAS
 
 | | HNE-dG adduct mismatched with dA in basic solution | | Descriptor: | (4S)-nonane-1,4-diol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DAP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-11-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational interconversion of the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N(2)-deoxyguanosine adduct when mismatched with deoxyadenosine in DNA
Chem.Res.Toxicol., 22, 2009
|
|
4ICZ
 
 | | HER2 1221/1222 phosphorylation regulated by PTPN9 | | Descriptor: | HER2, Tyrosine-protein phosphatase non-receptor type 9 | | Authors: | Xu, Y.F, Wang, H.M, Liu, C.H, Yu, X, Sun, J.P. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HER2 1221/1222 phosphorylation regulated by PTPN9
To be Published
|
|
7YFS
 
 | | The NMR structure of noursin, a tricyclic ribosomal peptide containing a histidine-to-butyrine crosslink | | Descriptor: | noursin | | Authors: | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
2LBI
 
 | | N2-dG:N2-dG interstrand cross-link induced by trans-4-hydroxynonenal | | Descriptor: | (4S)-nonane-1,4-diol, DNA (5'-D(*GP*CP*TP*AP*GP*CP*GP*AP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*CP*GP*CP*TP*AP*GP*C)-3') | | Authors: | Huang, H, Kozekov, I.D, Wang, H, Kozekova, A, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Formation of a N2-dG:N2-dG carbinolamine DNA cross-link by the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N2-dG adduct.
J.Am.Chem.Soc., 133, 2011
|
|
4GGV
 
 | | Crystal Structure of HmtT Involved in Himastatin Biosynthesis | | Descriptor: | Cytochrome P450 superfamily protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhang, H, Chen, J, Wang, H, Zhang, H. | | Deposit date: | 2012-08-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.331 Å) | | Cite: | Structural analysis of HmtT and HmtN involved in the tailoring steps of himastatin biosynthesis
Febs Lett., 587, 2013
|
|
3P3Q
 
 | | Crystal Structure of MmoQ Response regulator from Methylococcus capsulatus str. Bath at the resolution 2.4A, Northeast Structural Genomics Consortium Target McR175M | | Descriptor: | MmoQ, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Abashidze, M, Seetharaman, J, Janjua, J, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-10-05 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Northeast Structural Genomics Consortium Target McR175M
To be Published
|
|
5GW4
 
 | | Structure of Yeast NPP-TRiC | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
8HJE
 
 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | Descriptor: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG4
 
 | | Cryo-EM structure of nauclefine-induced PDE3A-SLFN12 complex | | Descriptor: | MAGNESIUM ION, Parvine, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
5GW5
 
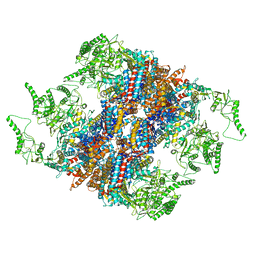 | | Structure of TRiC-AMP-PNP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
8HXX
 
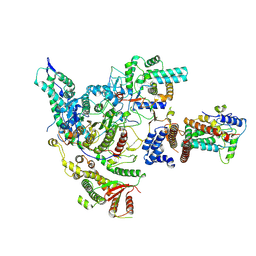 | | Cryo-EM structure of the histone deacetylase complex Rpd3S | | Descriptor: | Chromatin modification-related protein EAF3, Histone H3, Histone deacetylase RPD3, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HXZ
 
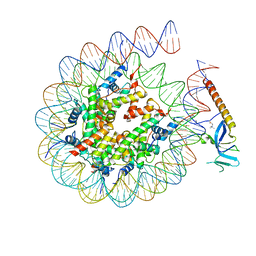 | | Cryo-EM structure of Eaf3 CHD in complex with nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (352-MER), Histone H2A, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
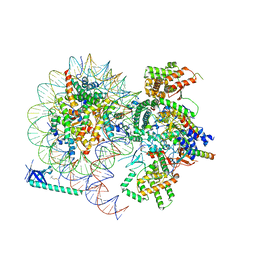
8HXY
 
 | |
7FH6
 
 | | Friedel-Crafts alkylation enzyme CylK | | Descriptor: | CALCIUM ION, CHLORIDE ION, CylK, ... | | Authors: | Liu, L, Wang, H.Q, Xiang, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
6DLU
 
 | | Cryo-EM of the GMPPCP-bound human dynamin-1 polymer assembled on the membrane in the constricted state | | Descriptor: | Dynamin-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
8HY0
 
 | |
7CXM
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
