3RD6
 
 | | Crystal structure of Mll3558 protein from Rhizobium loti. Northeast Structural Genomics Consortium target id MlR403 | | 分子名称: | Mll3558 protein | | 著者 | Seetharaman, J, Chen, Y, Wang, D, Ciccosanti, C, Sahdev, S, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-04-01 | | 公開日 | 2011-05-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of Mll3558 protein from Rhizobium loti. Northeast Structural Genomics Consortium target id MlR403
To be Published
|
|
3RD4
 
 | | Crystal structure of PROPEN_03304 from Proteus penneri ATCC 35198 Northeast Structural Genomics Consortium target id PvR55 | | 分子名称: | uncharacterized protein | | 著者 | Seetharaman, J, Min, S, Kuzin, A, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-03-31 | | 公開日 | 2011-05-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of PROPEN_03304 from Proteus penneri ATCC 35198 Northeast Structural Genomics Consortium target id PvR55
To be Published
|
|
4IOH
 
 | | Crystal Structure of the Tll1086 protein from Thermosynechococcus elongatus, Northeast Structural Genomics Consortium Target TeR258 | | 分子名称: | Tll1086 protein | | 著者 | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, X, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-01-07 | | 公開日 | 2013-01-30 | | 最終更新日 | 2013-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Crystal Structure of the Tll1086 protein from Thermosynechococcus elongatus.
To be Published
|
|
4INA
 
 | | Crystal Structure of the Q7MSS8_WOLSU protein from Wolinella succinogenes. Northeast Structural Genomics Consortium Target WsR35 | | 分子名称: | saccharopine dehydrogenase | | 著者 | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-01-04 | | 公開日 | 2013-01-16 | | 最終更新日 | 2013-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.488 Å) | | 主引用文献 | Crystal Structure of the Q7MSS8_WOLSU protein from Wolinella succinogenes.
To be Published
|
|
4MO1
 
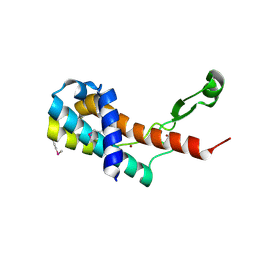 | | Crystal structure of antitermination protein Q from bacteriophage lambda. Northeast Structural Genomics Consortium target OR18A. | | 分子名称: | Antitermination protein Q, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Vorobiev, S, Su, M, Nickels, B, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Ebright, R.H, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-09-11 | | 公開日 | 2013-09-25 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Crystal structure of antitermination protein Q from bacteriophage lambda.
To be Published
|
|
4N9X
 
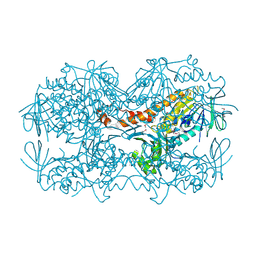 | | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161 | | 分子名称: | Putative monooxygenase | | 著者 | Kuzin, A, Chen, Y, Lew, S, Seetharaman, J, Mao, L, Xiao, R, Owens, L.A, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-10-21 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.503 Å) | | 主引用文献 | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161
To be Published
|
|
5YPI
 
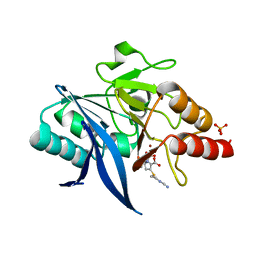 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI1 complex | | 分子名称: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Feng, H, Wang, D, Liu, W. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPK
 
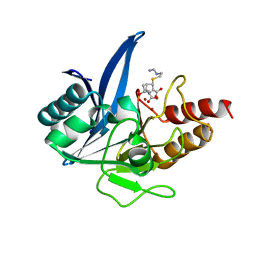 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI2 complex | | 分子名称: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | 著者 | Feng, H, Wang, D, Liu, W. | | 登録日 | 2017-11-02 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPN
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI2 complex | | 分子名称: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | 著者 | Feng, H, Liu, W, Wang, D. | | 登録日 | 2017-11-02 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPL
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EP complex | | 分子名称: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | 著者 | Feng, H, Wang, D, Liu, W. | | 登録日 | 2017-11-02 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPM
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI1 complex | | 分子名称: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | 著者 | Feng, H, Wang, D, Liu, W. | | 登録日 | 2017-11-02 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
4Q28
 
 | | Crystal Structure of the Plectin 1 and 2 Repeats of the Human Periplakin. Northeast Structural Genomics Consortium (NESG) Target HR9083A | | 分子名称: | Periplakin | | 著者 | Vorobiev, S, Lew, S, Seetharaman, J, Janjua, H, Xiao, R, O'Connell, P.T, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-04-07 | | 公開日 | 2014-06-18 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Crystal Structure of the Plectin 1 and 2 Repeats of Human Periplakin.
To be Published
|
|
7EPB
 
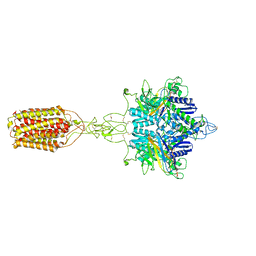 | | Cryo-EM structure of LY354740-bound mGlu2 homodimer | | 分子名称: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, Anti-RON nanobody, Metabotropic glutamate receptor 2 | | 著者 | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPC
 
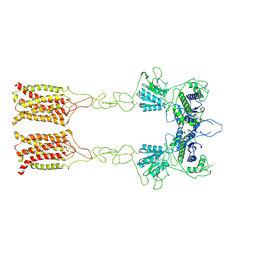 | | Cryo-EM structure of inactive mGlu7 homodimer | | 分子名称: | Isoform 3 of Metabotropic glutamate receptor 7 | | 著者 | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPA
 
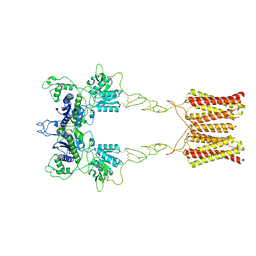 | | Cryo-EM structure of inactive mGlu2 homodimer | | 分子名称: | Metabotropic glutamate receptor 2 | | 著者 | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPD
 
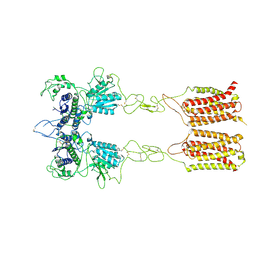 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | 分子名称: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | 著者 | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPF
 
 | | Crystal structure of mGlu2 bound to NAM597 | | 分子名称: | (8~{R})-4-[2,4-bis(fluoranyl)phenyl]-8-methyl-7-[(2-methylpyrazol-3-yl)methyl]-6,8-dihydro-5~{H}-1,7-naphthyridine-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | 著者 | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPE
 
 | | Crystal structure of mGlu2 bound to NAM563 | | 分子名称: | 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | 著者 | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | 登録日 | 2021-04-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7OR0
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 2 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P. | | 登録日 | 2021-06-04 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
To Be Published
|
|
6JRL
 
 | | Crystal structure of Drosophila alpha methyldopa-resistant protein/3,4-dihydroxyphenylacetaldehyde synthase | | 分子名称: | 3,4-dihydroxyphenylacetaldehyde synthase | | 著者 | Wei, S, Vavrick, C.J, Guan, H, Liao, C, Robinson, H, Liang, J, Wang, D, Han, Q, Li, J. | | 登録日 | 2019-04-04 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for the bifunctional mechanism of Drosophila alpha methyldopa-resistant protein/3,4-dihydroxyphenylacetaldehyde synthase
To Be Published
|
|
6KMJ
 
 | | Crystal structure of Sth1 bromodomain in complex with H3K14Ac | | 分子名称: | GLYCEROL, Histone H3, Nuclear protein STH1/NPS1 | | 著者 | Chen, G, Li, W, Yan, F, Wang, D, Chen, Y. | | 登録日 | 2019-07-31 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex.
Structure, 28, 2020
|
|
6KMB
 
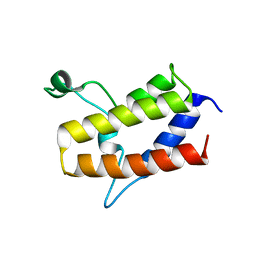 | | Crystal structure of Sth1 bromodomain | | 分子名称: | GLYCEROL, Nuclear protein STH1/NPS1 | | 著者 | Chen, G, Li, W, Yan, F, Wang, D, Chen, Y. | | 登録日 | 2019-07-31 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex.
Structure, 28, 2020
|
|
6L2N
 
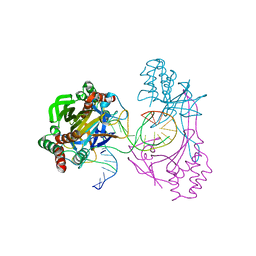 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(GTAC-3bp-GTAC) complex | | 分子名称: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*GP*TP*AP*CP*TP*AP*AP*GP*TP*AP*CP*TP*GP*CP*TP*GP*A)-3'), RE_R_Pab1 domain-containing protein | | 著者 | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | 登録日 | 2019-10-05 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
6L2O
 
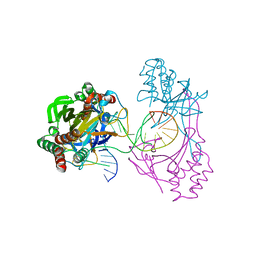 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(GTAC-5bp-GTAC) complex | | 分子名称: | DNA (5'-D(*CP*A*GP*CP*AP*GP*TP*AP*CP*TP*TP*AP*AP*AP*GP*TP*AP*CP*TP*GP*CP*TP*G)-3'), RE_R_Pab1 domain-containing protein | | 著者 | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | 登録日 | 2019-10-05 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
6M3L
 
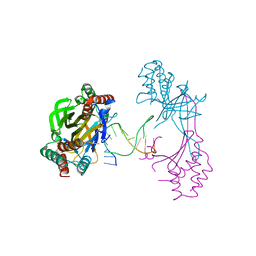 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(nonspecific) complex | | 分子名称: | DNA (5'-D(*CP*GP*CP*AP*TP*CP*GP*AP*TP*TP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*GP*CP*G)-3'), RE_R_Pab1 domain-containing protein | | 著者 | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | 登録日 | 2020-03-04 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
