8DH1
 
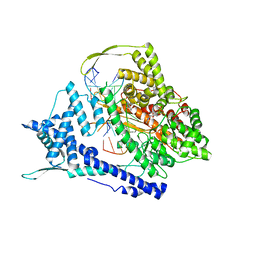 | | T7 RNA polymerase elongation complex with unnatural base dDs-PaTP pair | | Descriptor: | 1-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-ribofuranosyl}-1H-pyrrole-2-carbaldehyde, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2022-06-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of transcription recognition of a hydrophobic unnatural base pair by T7 RNA polymerase.
Nat Commun, 14, 2023
|
|
3ON9
 
 | | The SECRET domain from Ectromelia virus | | Descriptor: | Tumour necrosis factor receptor | | Authors: | Wang, X.Q, Xue, X.G, Wang, D.L. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis of chemokine sequestration by CrmD, a poxvirus-encoded tumor necrosis factor receptor
Plos Pathog., 7, 2011
|
|
4YAY
 
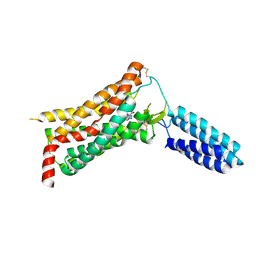 | | XFEL structure of human Angiotensin Receptor | | Descriptor: | 5,7-diethyl-1-{[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl}-3,4-dihydro-1,6-naphthyridin-2(1H)-one, Soluble cytochrome b562,Type-1 angiotensin II receptor | | Authors: | Zhang, H, Unal, H, Gati, C, Han, G.W, Zatsepin, N.A, James, D, Wang, D, Nelson, G, Weierstall, U, Messerschmidt, M, Williams, G.J, Boutet, S, Yefanov, O.M, White, T.A, Liu, W, Ishchenko, A, Tirupula, K.C, Desnoyer, R, Sawaya, M.C, Xu, Q, Coe, J, Cornrad, C.E, Fromme, P, Stevens, R.C, Katritch, V, Karnik, S.S, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Angiotensin receptor revealed by serial femtosecond crystallography.
Cell, 161, 2015
|
|
6EKK
 
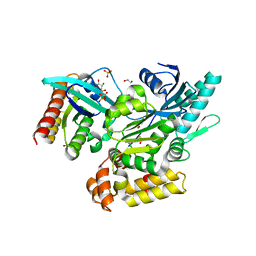 | | Crystal structure of GEF domain of DENND 1A in complex with Rab GTPase Rab35-GDP bound state. | | Descriptor: | 1,2-ETHANEDIOL, DENN domain-containing protein 1A, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Srikannathasan, V, Szykowska, A, Tallant, C, Strain-Damerell, C, Kopec, J, Kupinska, K, Mukhopadhyay, S, Gavin, M, Wang, D, Chalk, R, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Huber, K. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of DENND1A-RAB35 complex with GDP bound state.
To be published
|
|
8BLP
 
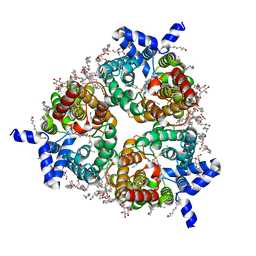 | | Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14 | | Descriptor: | 10-(4-ethylphenyl)sulfonyl-~{N}-(thiophen-2-ylmethyl)-5-thia-1,8,11,12-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,7,9,11-pentaen-7-amine, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Chi, G, Dietz, L, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, Scacioc, A, McKinley, G, Arrowsmith, C.H, Edwards, A, Bountra, C, Fernandez-Cid, A, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
7T9F
 
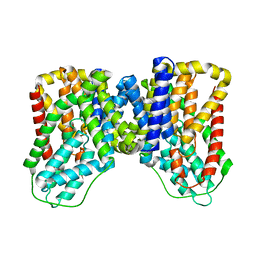 | | Structure of VcINDY-apo | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Song, J.M, Wang, D.N. | | Deposit date: | 2021-12-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of ion - substrate coupling in the Na + -dependent dicarboxylate transporter VcINDY.
Nat Commun, 13, 2022
|
|
7T9G
 
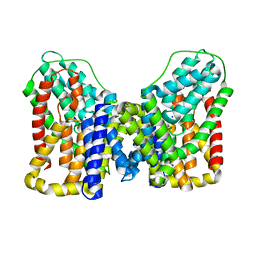 | | Structure of VcINDY-Na+ | | Descriptor: | DASS family sodium-coupled anion symporter, SODIUM ION | | Authors: | Sauer, D.B, Marden, J.J, Song, J.M, Wang, D.N. | | Deposit date: | 2021-12-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis of ion - substrate coupling in the Na + -dependent dicarboxylate transporter VcINDY.
Nat Commun, 13, 2022
|
|
6QYM
 
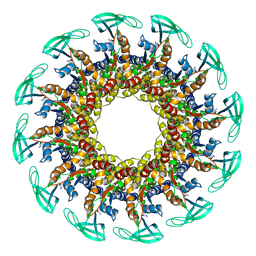 | |
4JLW
 
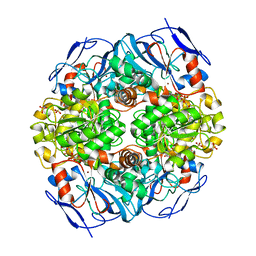 | | Crystal structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Glutathione-independent formaldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Chen, S, Liao, Y.P, Wang, D.L, Wang, S, Ding, J.F, Wang, Y.M, Cai, L.J, Ran, X.Y, Zhu, H.X. | | Deposit date: | 2013-03-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa: the binary complex with the cofactor NAD+.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6QYJ
 
 | |
5MR1
 
 | | Crystal structure of the Pleckstrin homology domain of Interactor protein for cytohesin exchange factors 1 (IPCEF1) | | Descriptor: | Interactor protein for cytohesin exchange factors 1 | | Authors: | Newman, J.A, Aitkenhead, H, Wang, D, Burgess-Brown, N, Williams, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the Pleckstrin homology domain of Interactor protein for cytohesin exchange factors 1 (IPCEF1)
To be published
|
|
3ONA
 
 | | The SECRET domain in complex with CX3CL1 | | Descriptor: | CX3CL1 protein, Tumour necrosis factor receptor | | Authors: | Wang, X.Q, Xue, X.G, Wang, D.L. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of chemokine sequestration by CrmD, a poxvirus-encoded tumor necrosis factor receptor
Plos Pathog., 7, 2011
|
|
2DSM
 
 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
6TTK
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
4Y7N
 
 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4UYI
 
 | | Crystal structure of the BTB domain of human SLX4 (BTBD12) | | Descriptor: | STRUCTURE-SPECIFIC ENDONUCLEASE SUBUNIT SLX4 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Strain-Damerell, C, Fairhead, M, Wang, D, Tallant, C, Cooper, C.D.O, Sorrell, F.J, Kopec, J, Chaikuad, A, Fitzpatrick, F, Pike, A.C.W, Hozjan, V, Ying, Z, Roos, A.K, Savitsky, P, Bradley, A, Nowak, R, Filippakopoulos, P, Krojer, T, Burgess-Brown, N.A, Marsden, B.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-09-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Slx4 (Btbd12)
To be Published
|
|
4Y52
 
 | | Crystal structure of 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4RL2
 
 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | (1R)-2-({(R)-carboxy[(2R,5S)-4-carboxy-5-methyl-5,6-dihydro-2H-1,3-thiazin-2-yl]methyl}amino)-2-oxo-1-phenylethanaminium, Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
4RM5
 
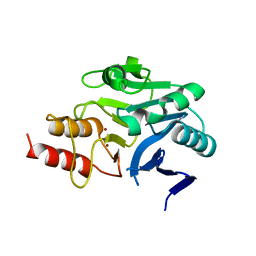 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
4RL0
 
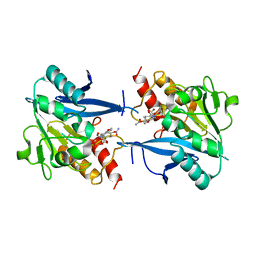 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
8EG6
 
 | | huCaspase-6 in complex with inhibitor 2a | | Descriptor: | (3R)-1-(ethanesulfonyl)-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
8EG5
 
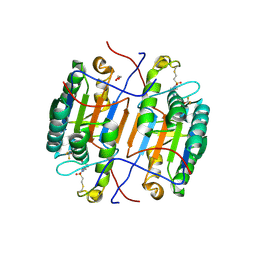 | | huCaspase-6 in complex with inhibitor 3a | | Descriptor: | (3R,5S)-1-(ethanesulfonyl)-5-phenyl-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide (bound form), 1,2-ETHANEDIOL, Caspase-6 subunit p11, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
4X31
 
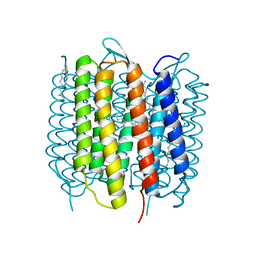 | | Room temperature structure of bacteriorhodopsin from lipidic cubic phase obtained with serial millisecond crystallography using synchrotron radiation | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, James, D, Wang, D, White, T, Zatsepin, N, Shilova, A, Nelson, G, Liu, H, Johansson, L, Heymann, M, Jaeger, K, Metz, M, Wickstrand, C, Wu, W, Baath, P, Berntsen, P, Oberthuer, D, Panneels, V, Cherezov, V, Chapman, H, Spence, J, Schertler, G, Neutze, R, Moraes, I, Burghammer, M, Standfuss, J, Weierstall, U. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipidic cubic phase serial millisecond crystallography using synchrotron radiation.
Iucrj, 2, 2015
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
5NXD
 
 | | LIM Domain Kinase 2 (LIMK2) In Complex With TH-300 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-chlorophenyl)sulfamoyl]-~{N}-(phenylmethyl)-~{N}-propyl-benzamide, LIM domain kinase 2 | | Authors: | Mathea, S, Salah, E, Hanke, T, Newman, J.A, Oerum, S, Wang, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Knapp, S. | | Deposit date: | 2017-05-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | LIM Domain Kinase 2 (LIMK2)In Complex With TH-300
To Be Published
|
|
