8SA3
 
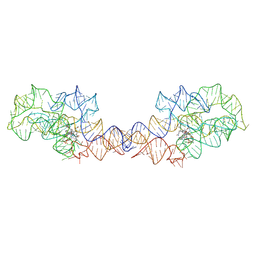 | | Adenosylcobalamin-bound riboswitch dimer, form 2 | | 分子名称: | Adenosylcobalamin, adenosylcobalamin riboswitch form 2 | | 著者 | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | 登録日 | 2023-03-31 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA6
 
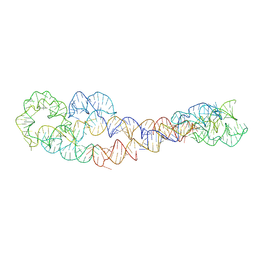 | | apo form of adenosylcobalamin riboswitch dimer | | 分子名称: | apo form of adenosylcobalamin riboswitch dimer | | 著者 | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | 登録日 | 2023-03-31 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
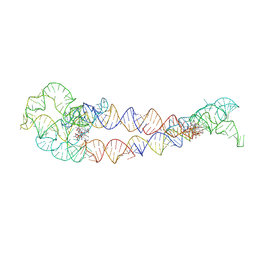 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | 分子名称: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | 著者 | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | 登録日 | 2023-03-31 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7CI1
 
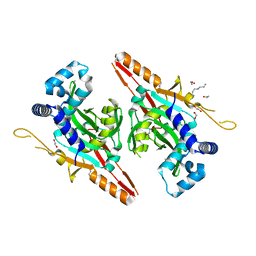 | |
4OBY
 
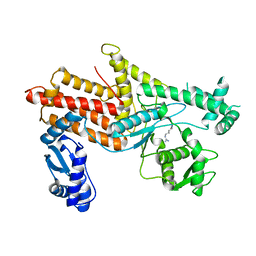 | | Crystal Structure of E.coli Arginyl-tRNA Synthetase and Ligand Binding Studies Revealed Key Residues in Arginine Recognition | | 分子名称: | ARGININE, Arginine--tRNA ligase | | 著者 | Bi, K, Zheng, Y, Dong, J, Gao, F, Wang, J, Wang, Y, Gong, W. | | 登録日 | 2014-01-08 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.574 Å) | | 主引用文献 | Crystal structure of E. coli arginyl-tRNA synthetase and ligand binding studies revealed key residues in arginine recognition.
Protein Cell, 5, 2014
|
|
8IU7
 
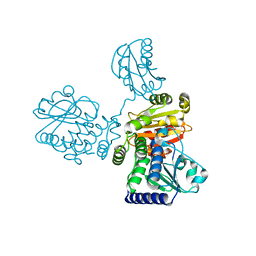 | |
3GO6
 
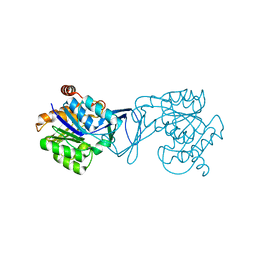 | | Crystal Structure of M. tuberculosis ribokinase (Rv2436) in complex with ribose and AMP-PNP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOKINASE RBSK, ... | | 著者 | Masters, E.I, Sun, Y, Wang, Y, Parker, W.B, Li, R. | | 登録日 | 2009-03-18 | | 公開日 | 2010-03-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structure and biochemical characterization of M.tuberculosis ribokinase (Rv2436)
To be Published
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | 分子名称: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | 著者 | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | 登録日 | 2019-06-27 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
3HHS
 
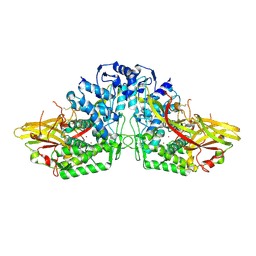 | | Crystal Structure of Manduca sexta prophenoloxidase | | 分子名称: | COPPER (II) ION, Phenoloxidase subunit 1, Phenoloxidase subunit 2 | | 著者 | Li, Y, Wang, Y, Jiang, H, Deng, J. | | 登録日 | 2009-05-17 | | 公開日 | 2009-09-29 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4NCA
 
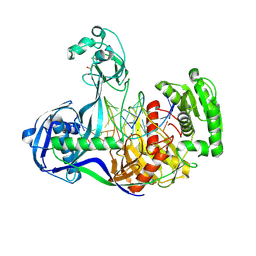 | | Structure of Thermus thermophilus Argonaute bound to guide DNA 19-mer and target DNA in the presence of Mg2+ | | 分子名称: | 5'-D(*AP*CP*AP*AP*CP*C)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-24 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.489 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5WTJ
 
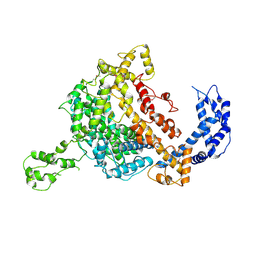 | | Crystal structure of an endonuclease | | 分子名称: | CRISPR-associated endoribonuclease C2c2 | | 著者 | Liu, L, Wang, Y. | | 登録日 | 2016-12-13 | | 公開日 | 2017-02-08 | | 最終更新日 | 2017-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.503 Å) | | 主引用文献 | Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities
Cell, 168, 2017
|
|
7UMS
 
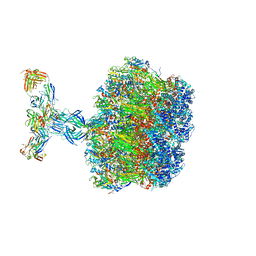 | | Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 in complex with antibody 41 - Upright conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fab 41 heavy chain, ... | | 著者 | Jenni, S, Zongli, L, Wang, Y, Bessey, T, Salgado, E.N, Schmidt, A.G, Greenberg, H.B, Jiang, B, Harrison, S.C. | | 登録日 | 2022-04-07 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.
J.Virol., 96, 2022
|
|
2AOC
 
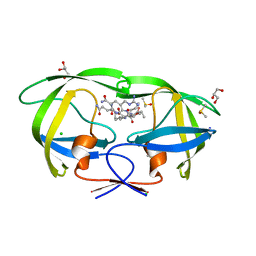 | | Crystal structure analysis of HIV-1 protease mutant I84V with a substrate analog P2-NC | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
7UMT
 
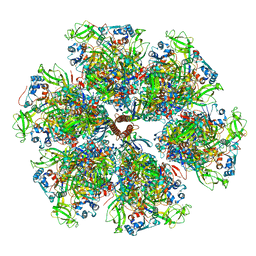 | | Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 - Reversed conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | 著者 | Jenni, S, Zongli, L, Wang, Y, Bessey, T, Salgado, E.N, Schmidt, A.G, Greenberg, H.B, Jiang, B, Harrison, S.C. | | 登録日 | 2022-04-07 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.
J.Virol., 96, 2022
|
|
2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | 分子名称: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOD
 
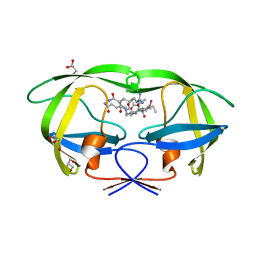 | | Crystal structure analysis of HIV-1 protease with a substrate analog P2-NC | | 分子名称: | DIMETHYL SULFOXIDE, GLYCEROL, HIV-1 PROTEASE, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
1YT3
 
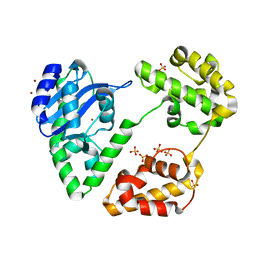 | | Crystal Structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing | | 分子名称: | Ribonuclease D, SULFATE ION, ZINC ION | | 著者 | Zuo, Y, Wang, Y, Malhotra, A. | | 登録日 | 2005-02-09 | | 公開日 | 2005-08-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of Escherichia coli RNase D, an Exoribonuclease Involved in Structured RNA Processing
Structure, 13, 2005
|
|
4N47
 
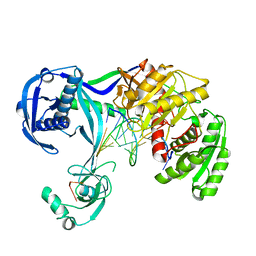 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 12-mer target DNA | | 分子名称: | 5'-D(*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-08 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.823 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6E6S
 
 | | 1.45 A resolution structure of the C-terminally truncated [2Fe-2S] ferredoxin (Bfd) R26E/K46Y mutant from Pseudomonas aeruginosa | | 分子名称: | Bacterioferritin-associated ferredoxin, FE2/S2 (INORGANIC) CLUSTER | | 著者 | Lovell, S, Wijerathne, H, Battaile, K.P, Yao, H, Wang, Y, Rivera, M. | | 登録日 | 2018-07-25 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site.
Biochemistry, 57, 2018
|
|
5XP8
 
 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 4A5 on the guide strand | | 分子名称: | DNA (5'-D(*AP*GP*T)-3'), DNA (5'-D(*CP*AP*AP*CP*C*AP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(P*TP*GP*AP*GP*AP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*T)-3'), ... | | 著者 | Sheng, G, Gogakos, T, Wang, J, Zhao, H, Serganov, A, Juranek, S, Tuschl, T, Patel, J.D, Wang, Y. | | 登録日 | 2017-06-01 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.
Nucleic Acids Res., 45, 2017
|
|
7DHJ
 
 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | 分子名称: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | 著者 | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | 登録日 | 2020-11-15 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.962 Å) | | 主引用文献 | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
7DQ9
 
 | |
6E6Q
 
 | | 1.20 A resolution structure of the C-terminally truncated [2Fe-2S] ferredoxin (Bfd) from Pseudomonas aeruginosa | | 分子名称: | Bacterioferritin-associated ferredoxin, FE2/S2 (INORGANIC) CLUSTER | | 著者 | Lovell, S, Wijerathne, H, Battaile, K.P, Yao, H, Wang, Y, Rivera, M. | | 登録日 | 2018-07-25 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site.
Biochemistry, 57, 2018
|
|
5XOW
 
 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 6'A7' on the target strand | | 分子名称: | DNA (5'-D(P*(TD)P*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), MAGNESIUM ION, RNA (5'-R(P*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*AP*CP*CP*UP*CP*G)-3'), ... | | 著者 | Sheng, G, Wang, J, Zhao, H, Wang, Y. | | 登録日 | 2017-05-31 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.902 Å) | | 主引用文献 | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes
Nucleic Acids Res., 45, 2017
|
|
4NCB
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 19-mer target DNA with Mg2+ | | 分子名称: | 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*C)-3', 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-24 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.189 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
