8ZC9
 
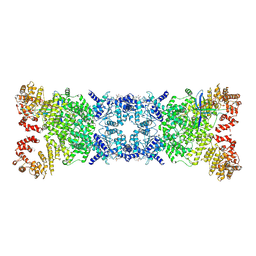 | | The Cryo-EM structure of DSR2-Tail tube-NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein, tail tube protein | | Authors: | Wang, R, Xu, Q, Wu, Z, Li, J, Yang, R, Shi, Z, Li, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
7QVK
 
 | | NM-02 in complex with HER2-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NM-02, Receptor tyrosine-protein kinase erbB-2, ... | | Authors: | Cowan, R, Hall, G, Carr, M. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Co-crystallisation and humanisation of an anti-HER2 single-domain antibody as a theranostic tool.
Plos One, 18, 2023
|
|
7KHR
 
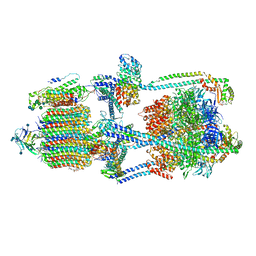 | | Cryo-EM structure of bafilomycin A1-bound intact V-ATPase from bovine brain | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (5R)-2,4-dideoxy-1-C-{(2S,3R,4S)-3-hydroxy-4-[(2R,3S,4E,6E,9R,10S,11R,12E,14Z)-10-hydroxy-3,15-dimethoxy-7,9,11,13-tetramethyl-16-oxo-1-oxacyclohexadeca-4,6,12,14-tetraen-2-yl]pentan-2-yl}-4-methyl-5-propan-2-yl-alpha-D-threo-pentopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-17 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Molecular basis of V-ATPase inhibition by bafilomycin A1.
Nat Commun, 12, 2021
|
|
7UNF
 
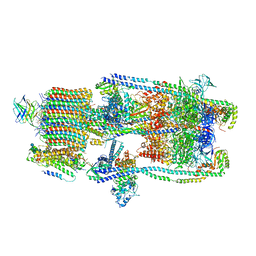 | | CryoEM structure of a mEAK7 bound human V-ATPase complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2022-04-10 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Molecular basis of mEAK7-mediated human V-ATPase regulation.
Nat Commun, 13, 2022
|
|
7UNE
 
 | |
6UM1
 
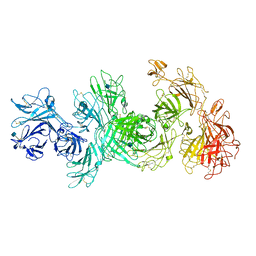 | | Structure of M-6-P/IGFII Receptor at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, ... | | Authors: | Wang, R, Qi, X, Li, X. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Sci Adv, 6, 2020
|
|
6UM2
 
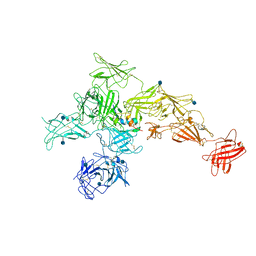 | | Structure of M-6-P/IGFII Receptor and IGFII complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, Insulin-like growth factor II | | Authors: | Wang, R, Qi, X, Li, X. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Sci Adv, 6, 2020
|
|
5H43
 
 | |
5GO0
 
 | |
3G5B
 
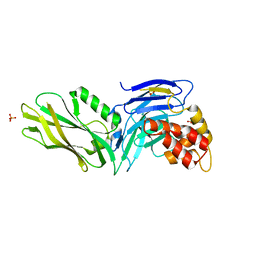 | | The structure of UNC5b cytoplasmic domain | | Descriptor: | Netrin receptor UNC5B, PHOSPHATE ION | | Authors: | Wang, R, Wei, Z, Zhang, M. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Autoinhibition of UNC5b revealed by the cytoplasmic domain structure of the receptor
Mol.Cell, 33, 2009
|
|
4U32
 
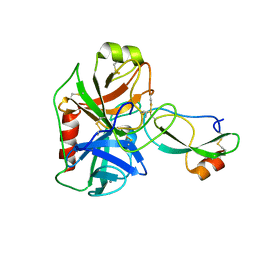 | | Human mesotrypsin complexed with HAI-2 Kunitz domain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Kunitz-type protease inhibitor 2, ... | | Authors: | Wang, R, Soares, A.S, Radisky, E.S. | | Deposit date: | 2014-07-18 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Sequence and Conformational Specificity in Substrate Recognition: SEVERAL HUMAN KUNITZ PROTEASE INHIBITOR DOMAINS ARE SPECIFIC SUBSTRATES OF MESOTRYPSIN.
J.Biol.Chem., 289, 2014
|
|
4U30
 
 | |
3GS2
 
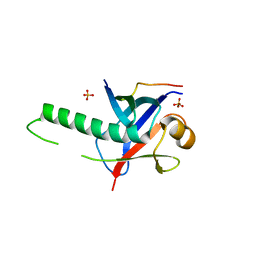 | | Ring1B C-terminal domain/Cbx7 Cbox Complex | | Descriptor: | Chromobox protein homolog 7, E3 ubiquitin-protein ligase RING2, SULFATE ION, ... | | Authors: | Wang, R, Taylor, A.B, Kim, C.A. | | Deposit date: | 2009-03-26 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Polycomb Group Targeting through Different Binding Partners of RING1B C-Terminal Domain.
Structure, 18, 2010
|
|
3IXS
 
 | | Ring1B C-terminal domain/RYBP C-terminal domain Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, E3 ubiquitin-protein ligase RING2, ... | | Authors: | Wang, R, Taylor, A.B, Kim, C.A. | | Deposit date: | 2009-09-04 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polycomb Group Targeting through Different Binding Partners of RING1B C-Terminal Domain.
Structure, 18, 2010
|
|
3E6F
 
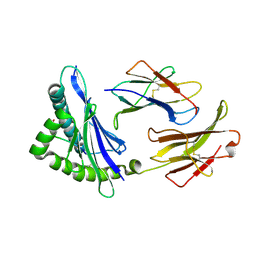 | | MHC CLASS I H-2Dd Heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PA9, from the Human immunodeficiency virus (BaL) envelope glycoprotein 120 | | Descriptor: | BETA-2 MICROGLOBULIN, Envelope glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
3E6H
 
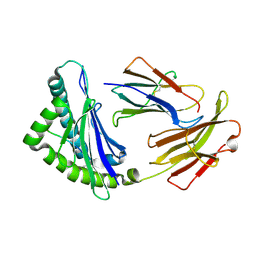 | | MHC CLASS I H-2Dd heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PI10, from the human immunodeficiency virus (BaL) envelope glycoprotein 120 | | Descriptor: | Envelope glycoprotein 10-residue peptide, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
4G59
 
 | |
3DMM
 
 | | Crystal structure of the CD8 alpha beta/H-2Dd complex | | Descriptor: | Beta-2 microglobulin, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Wang, R, Natarajan, K, Margulies, D.H. | | Deposit date: | 2008-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the CD8alphabeta/MHC class i interaction: focused recognition orients CD8beta to a T cell proximal position.
J.Immunol., 183, 2009
|
|
4FGY
 
 | | Identification of a unique PPAR ligand with an unexpected binding mode and antibetic activity | | Descriptor: | (4R,6S,8S,12R,14R,16Z,18R,19R,20S,21S)-19,21-dihydroxy-22-{(2S,2'R,5S,5'S)-5'-[(1R)-1-hydroxyethyl]-2,5'-dimethyloctahydro-2,2'-bifuran-5-yl}-4,6,8,12,14,18,20-heptamethyl-9,11-dioxodocos-16-enoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, R, Li, Y. | | Deposit date: | 2012-06-05 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Identification of the antibiotic ionomycin as an unexpected peroxisome proliferator-activated receptor Gamma (PPAR-gamma) ligand with a unique binding mode and effective glucose-lowering activity in a mouse model of diabetes.
Diabetologia, 56, 2013
|
|
4PUT
 
 | |
3PKM
 
 | | Crystal structure of Cas6 with its substrate RNA | | Descriptor: | 5'-R(*AP*UP*UP*AP*CP*AP*AP*UP*AP*A)-3', 5'-R(P*UP*UP*AP*CP*AP*AP*UP*AP*A)-3', CRISPR-associated endoribonuclease Cas6 | | Authors: | Wang, R, Preamplume, G, Li, H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Interaction of the Cas6 Riboendonuclease with CRISPR RNAs: Recognition and Cleavage.
Structure, 19, 2011
|
|
8BB7
 
 | |
3QJL
 
 | |
8B3K
 
 | |
8BF4
 
 | | Crystal structure of Mouse Plexin-B1 (20-535) in complex with VHH15 and VHH14 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Cowan, R, Hall, G, Carr, M. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nanobody inhibitors of Plexin-B1 identify allostery in plexin-semaphorin interactions and signaling.
J.Biol.Chem., 299, 2023
|
|
