7EG4
 
 | | Cryo-EM structure of nauclefine-induced PDE3A-SLFN12 complex | | Descriptor: | MAGNESIUM ION, Parvine, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | Descriptor: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
3NGW
 
 | | Crystal Structure of Molybdopterin-guanine dinucleotide biosynthesis protein A from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR189 | | Descriptor: | CALCIUM ION, Molybdopterin-guanine dinucleotide biosynthesis protein A (MobA) | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-13 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Northeast Structural Genomics Consortium Target GR189
To be Published
|
|
3ESH
 
 | | Crystal structure of a probable metal-dependent hydrolase from Staphylococcus aureus. Northeast Structural Genomics target ZR314 | | Descriptor: | Protein similar to metal-dependent hydrolase | | Authors: | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a probable metal-dependent hydrolase from
Staphylococcus aureus. Northeast Structural Genomics target ZR314
To be Published
|
|
9GG2
 
 | | Structure of IPNV L5 capsid | | Descriptor: | Structural polyprotein | | Authors: | Munke, A, Gamil, A, Mikalsen, A, Wang, H, Evensen, O, Okamoto, K. | | Deposit date: | 2024-08-13 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure of IPNV L5 capsid
To Be Published
|
|
3O6U
 
 | | Crystal Structure of CPE2226 protein from Clostridium perfringens. Northeast Structural Genomics Consortium Target CpR195 | | Descriptor: | uncharacterized protein CPE2226 | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of CPE2226 protein from Clostridium perfringens.
To be Published
|
|
3VPS
 
 | | Structure of a novel NAD dependent-NDP-hexosamine 5,6-dehydratase, TunA, involved in tunicamycin biosynthesis | | Descriptor: | NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Wyszynski, F.J, Lee, S.S, Yabe, T, Wang, H, Gomez-Escribano, J.P, Bibb, M.J, Lee, S.J, Davies, G.J, Davis, B.G. | | Deposit date: | 2012-03-12 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biosynthesis of nucleoside antibiotic tunicamycin proceeds via unique exo-glycal intermediates
To be published
|
|
9BJK
 
 | | Inactive mu opioid receptor bound to Nb6, naloxone and NAM | | Descriptor: | Mu-type opioid receptor, Naloxone, Nalpha-[({(1M)-1-[5-(benzyloxy)pyridin-3-yl]naphthalen-2-yl}sulfanyl)acetyl]-3-methoxy-N,4-dimethyl-L-phenylalaninamide, ... | | Authors: | O'Brien, E.S, Wang, H, Kaavya Krishna, K, Zhang, C, Kobilka, B.K. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | A mu-opioid receptor modulator that works cooperatively with naloxone.
Nature, 631, 2024
|
|
8HJE
 
 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
6KE4
 
 | | ABloop reengineered Ferritin Nanocage | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Wang, W.M, Wang, H.F. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AB loop engineered ferritin nanocages for drug loading under benign experimental conditions.
Chem.Commun.(Camb.), 55, 2019
|
|
3NV5
 
 | | Crystal Structure of Cytochrome P450 CYP101D2 | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
4LH6
 
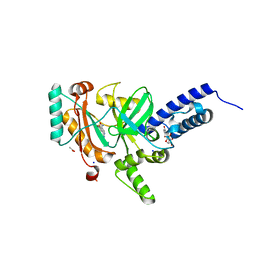 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-amino-2-bromothieno[3,2-c]pyridine-7-carboxamide, ACETATE ION, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ... | | Authors: | Benenato, K, Wang, H, Mcguire, H.M, Davis, H, Gao, N, Prince, D.B, Jahic, H, Stokes, S.S, Boriack-Sjodin, P.A. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7FH6
 
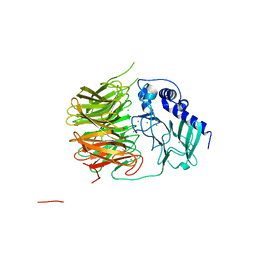 | | Friedel-Crafts alkylation enzyme CylK | | Descriptor: | CALCIUM ION, CHLORIDE ION, CylK, ... | | Authors: | Liu, L, Wang, H.Q, Xiang, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
8JWH
 
 | |
5WH6
 
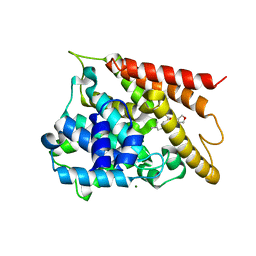 | | Crystal structure of PDE4D2 in complex with inhibitor (S_Zl-n-91) | | Descriptor: | 1-[4-(difluoromethoxy)-3-{[(3S)-oxolan-3-yl]oxy}phenyl]-3-methylbutan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H, Wang, H. | | Deposit date: | 2017-07-14 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a PDE4-Specific Pocket for the Design of Selective Inhibitors.
Biochemistry, 57, 2018
|
|
3CEW
 
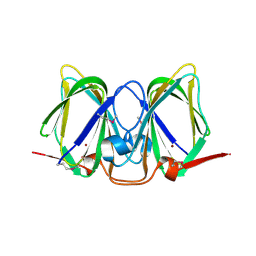 | | Crystal structure of a cupin protein (BF4112) from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR205 | | Descriptor: | Uncharacterized cupin protein, ZINC ION | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Wang, H, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of a cupin protein (BF4112) from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR205.
To be Published
|
|
3FOJ
 
 | | Crystal Structure of SSP1007 From Staphylococcus saprophyticus subsp. saprophyticus. Northeast Structural Genomics Target SyR101A. | | Descriptor: | SODIUM ION, uncharacterized protein | | Authors: | Seetharaman, J, Abashidze, M, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-30 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of SSP1007 From Staphylococcus saprophyticus subsp. saprophyticus. Northeast Structural Genomics Target SyR101A.
To be Published
|
|
8XWB
 
 | | Crystal structure of dinitrosyl iron units binding with human heavy chain Ferritin | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin heavy chain, ... | | Authors: | Gong, W.J, Wang, W.M, Wang, H.F. | | Deposit date: | 2024-01-16 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Insight into the photodynamic mechanism and protein binding of a nitrosyl iron-sulfur [Fe 2 S 2 (NO) 4 ] 2- cluster.
Spectrochim Acta A Mol Biomol Spectrosc, 320, 2024
|
|
3MCU
 
 | | Crystal structure of the dipicolinate synthase chain B from Bacillus cereus. Northeast Structural Genomics Consortium Target BcR215. | | Descriptor: | Dipicolinate synthase, B chain, PHOSPHATE ION | | Authors: | Vorobiev, S, Lew, S, Abashidze, M, Seetharaman, J, Wang, H, Ciccosanti, C, Foote, E.L, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Crystal structure of the dipicolinate synthase chain B from Bacillus cereus.
To be Published
|
|
5YZ0
 
 | | Cryo-EM Structure of human ATR-ATRIP complex | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR | | Authors: | Rao, Q, Liu, M, Tian, Y, Wu, Z, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of human ATR-ATRIP complex.
Cell Res., 28, 2018
|
|
3E0E
 
 | | Crystal structure of a domain of replication protein A from Methanococcus maripaludis. NorthEast Structural Genomics targe MrR110B | | Descriptor: | Replication protein A | | Authors: | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a domain of replication protein A from Methanococcus maripaludis. NorthEast Structural Genomics targe MrR110B
To be Published
|
|
8Y8C
 
 | | Structure of HCoV-HKU1C spike in the inactive-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8E
 
 | | Structure of HCoV-HKU1C spike in the inactive-2up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y87
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y89
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Wang, H.F, Zhang, X, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
