1NQV
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-nitroso-6-ribityl-amino-2,4(1H,3H)pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
4OMR
 
 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX in complex with acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, BENZAMIDINE, Enoyl-CoA hydratase | | Authors: | Langner, K.M, Cooper, D.R, Chapman, H.C, Handing, K.B, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-27 | | Release date: | 2014-04-02 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX in complex with acetoacetyl-CoA
To be Published
|
|
3OQM
 
 | | structure of ccpa-hpr-ser46p-ackA2 complex | | Descriptor: | 5'-D(*TP*TP*GP*AP*TP*AP*AP*CP*GP*CP*TP*TP*AP*CP*AP*A)-3', 5'-D(*TP*TP*GP*TP*AP*AP*GP*CP*GP*TP*TP*AP*TP*CP*AP*A)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
1NTH
 
 | | Crystal structure of the methanosarcina barkeri monomethylamine methyltransferase (MTMB) | | Descriptor: | Monomethylamine methyltransferase mtmB1 | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase
Science, 296, 2002
|
|
5VNE
 
 | |
3ORQ
 
 | | Crystal Structure of N5-Carboxyaminoimidazole synthetase from Staphylococcus aureus complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4PBV
 
 | | Crystal structure of chicken receptor protein tyrosine phosphatase sigma in complex with TrkC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NT-3 growth factor receptor, Protein-tyrosine phosphatase CRYPalpha1 isoform, ... | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
4P0Y
 
 | |
4AEL
 
 | | PDE10A in complex with the inhibitor AZ5 | | Descriptor: | 2-(2'-ETHOXYBIPHENYL-4-YL)-4-HYDROXY-1,6-NAPHTHYRIDINE-3-CARBONITRILE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Bauer, U, Giordanetto, F, Bauer, M, OMahony, G, Johansson, K.E, Knecht, W, Hartleib-Geschwindner, J, Toppner Carlsson, E, Enroth, C, Sjogren, T. | | Deposit date: | 2012-01-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of 4-Hydroxy-1,6-Naphthyridine-3-Carbonitrile Derivatives as Novel Pde10A Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5VPG
 
 | | CRYSTAL STRUCTURE OF DER P 1 COMPLEXED WITH FAB 4C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
5YZ4
 
 | | Structure of the PIN domain endonuclease Utp24 | | Descriptor: | CALCIUM ION, ZINC ION, rRNA-processing protein fcf1 | | Authors: | Du, Y, An, W, Ye, K. | | Deposit date: | 2017-12-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Structural and functional analysis of Utp24, an endonuclease for processing 18S ribosomal RNA.
Plos One, 13, 2018
|
|
5Z08
 
 | |
4P5A
 
 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | Descriptor: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
5Z2S
 
 | | Crystal structure of DUX4-HD2 domain | | Descriptor: | Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
1L7N
 
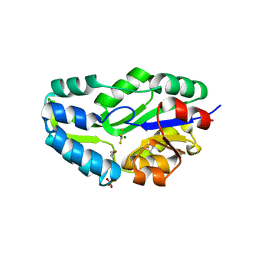 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
3OT1
 
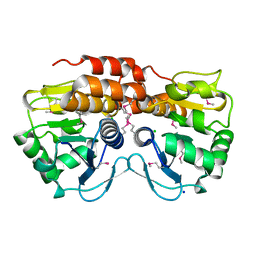 | | Crystal structure of VC2308 protein | | Descriptor: | 4-methyl-5(B-hydroxyethyl)-thiazole monophosphate biosynthesis enzyme, CHLORIDE ION, SODIUM ION | | Authors: | Niedzialkowska, E, Wawrzak, Z, Chruszcz, M, Porebski, P, Skarina, T, Huang, X, Grimshaw, S, Cymborowski, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of VC2308 protein
To be Published
|
|
5VG6
 
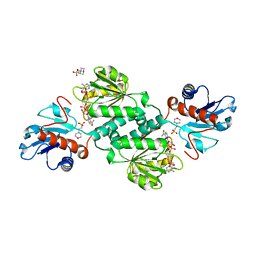 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lipowska, J, Shabalin, I.G, Kutner, J, Gasiorowska, O.A, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-04-10 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 in complex with NADPH and MES.
to be published
|
|
4PMO
 
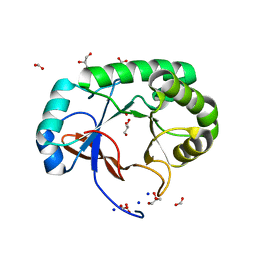 | | Crystal structure of the Mycobacterium tuberculosis Tat-secreted protein Rv2525c, monoclinic crystal form I | | Descriptor: | FORMIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Bellinzoni, M, Haouz, A, Shepard, W, Alzari, P.M. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural studies suggest a peptidoglycan hydrolase function for the Mycobacterium tuberculosis Tat-secreted protein Rv2525c.
J.Struct.Biol., 188, 2014
|
|
1LF7
 
 | | Crystal Structure of Human Complement Protein C8gamma at 1.2 A Resolution | | Descriptor: | CITRIC ACID, Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
5ZHI
 
 | | Apo crystal structure of TrmD from Mycobacterium tuberculosis | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5VR6
 
 | | Structure of Human Sts-1 histidine phosphatase domain with sulfate bound | | Descriptor: | SULFATE ION, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Zhou, W, Yin, Y, Weinheimer, A.W, Kaur, N, Carpino, N, French, J.B. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Biochemistry, 56, 2017
|
|
5ZHM
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-({4-[(diethylamino)methyl]phenyl}methyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
4AQE
 
 | | CRYSTAL STRUCTURE OF DEAFNESS ASSOCIATED MUTANT MOUSE CADHERIN-23 EC1- 2S70P AND PROTOCADHERIN-15 EC1-2 FORM I | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CADHERIN-23, CALCIUM ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2012-04-16 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Force-Conveying Cadherin Bond Essential for Inner-Ear Mechanotransduction
Nature, 492, 2012
|
|
1L2Q
 
 | | Crystal Structure of the Methanosarcina barkeri Monomethylamine Methyltransferase (MtmB) | | Descriptor: | AMMONIUM ION, monomethylamine methyltransferase | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2002-02-24 | | Release date: | 2002-06-05 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase.
Science, 296, 2002
|
|
4A99
 
 | | STRUCTURE OF THE TETRACYCLINE DEGRADING MONOOXYGENASE TETX IN COMPLEX WITH MINOCYCLINE | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | Deposit date: | 2011-11-25 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Putative Dioxygen-Binding Sites and Recognition of Tigecycline and Minocycline in the Tetracycline-Degrading Monooxygenase Tetx
Acta Crystallogr.,Sect.D, 69, 2013
|
|
