3RPL
 
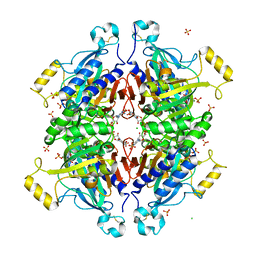 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 in complex with FRUCTOSE-1,6-BISPHOSPHATE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
5WB9
 
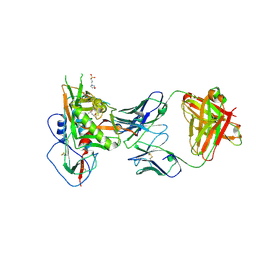 | | Crystal structure of CD4 binding site antibody N60P23 in complex with HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Gohain, N, Tolbert, W, Pazgier, M. | | Deposit date: | 2017-06-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Near-Pan-neutralizing Antibodies against HIV-1 by Deconvolution of Plasma Humoral Responses.
Cell, 173, 2018
|
|
4PVH
 
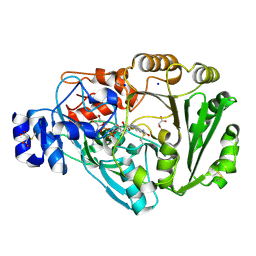 | | Phl p 4 N158H variant, a glucose dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Pollen allergen Phl p 4.0202, ... | | Authors: | Zafred, D, Teufelberger, A, Keller, W, Macheroux, P. | | Deposit date: | 2014-03-17 | | Release date: | 2014-04-02 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
5YBF
 
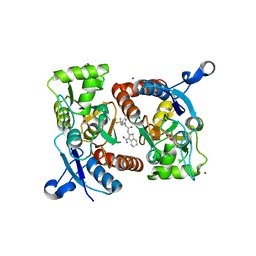 | | Crystal structure of the GluA2o LBD in complex with glutamate and HBT1 | | Descriptor: | 2-[2-[5-methyl-3-(trifluoromethyl)pyrazol-1-yl]ethanoylamino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | HBT1, a Novel AMPA Receptor Potentiator with Lower Agonistic Effect, Avoided Bell-Shaped Response in In Vitro BDNF Production.
J. Pharmacol. Exp. Ther., 364, 2018
|
|
4PZI
 
 | | Zinc finger region of MLL2 in complex with CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3'), Histone-lysine N-methyltransferase 2B, UNKNOWN ATOM OR ION, ... | | Authors: | Chao, X, Tempel, W, Liu, K, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
3ZW1
 
 | | Structure of Bambl lectine in complex with lewix x antigen | | Descriptor: | BAMBL LECTIN, alpha-D-galactopyranose, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Audfray, A, Claudinon, J, Abounit, S, Ruvoen-Clouet, N, Larson, G, Wimmerova, M, Lependu, J, Romer, W, Varrot, A, Imberty, A. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deciphering the Glycan Preference of Bacterial Lectins by Glycan Array and Molecular Docking with Validation by Microcalorimetry and Crystallography.
Plos One, 8, 2013
|
|
1COS
 
 | | CRYSTAL STRUCTURE OF A SYNTHETIC TRIPLE-STRANDED ALPHA-HELICAL BUNDLE | | Descriptor: | COILED SERINE | | Authors: | Lovejoy, B, Choe, S, Cascio, D, Mcrorie, D.K, Degrado, W, Eisenberg, D. | | Deposit date: | 1993-01-22 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a synthetic triple-stranded alpha-helical bundle.
Science, 259, 1993
|
|
1P6C
 
 | | crystal structure of phosphotriesterase triple mutant H254G/H257W/L303T complexed with diisopropylmethylphosphonate | | Descriptor: | DIETHYL 4-METHYLBENZYLPHOSPHONATE, METHYLPHOSPHONIC ACID DIISOPROPYL ESTER, Parathion hydrolase, ... | | Authors: | Hill, C.M, Li, W, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2003-04-29 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhanced degradation of chemical warfare agents through molecular engineering of the phosphotriesterase active site.
J.Am.Chem.Soc., 125, 2003
|
|
3ROJ
 
 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
4MYM
 
 | | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, AL Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-27 | | Release date: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides.
To be Published
|
|
6AAA
 
 | |
4PWB
 
 | | Phl p 4 I153V variant, a glucose oxidase, pressurized with Xenon | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Pollen allergen Phl p 4.0202, ... | | Authors: | Zafred, D, Keller, W, Macheroux, P. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
5V19
 
 | |
1FHG
 
 | | HIGH RESOLUTION REFINEMENT OF TELOKIN | | Descriptor: | TELOKIN | | Authors: | Tomchick, D.R, Minor, W, Kiyatkin, A, Lewinski, K, Somlyo, A.V, Somlyo, A.P. | | Deposit date: | 2000-08-01 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
1BVR
 
 | | M.TB. ENOYL-ACP REDUCTASE (INHA) IN COMPLEX WITH NAD+ AND C16-FATTY-ACYL-SUBSTRATE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE), TRANS-2-HEXADECENOYL-(N-ACETYL-CYSTEAMINE)-THIOESTER | | Authors: | Rozwarski, D.A, Vilcheze, C, Sugantino, M, Bittman, R, Jacobs, W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate.
J.Biol.Chem., 274, 1999
|
|
4COO
 
 | | Crystal structure of human cystathionine beta-synthase (delta516-525) at 2.0 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, ... | | Authors: | McCorvie, T.J, Kopec, J, Vollamar, M, Strain-Damerell, C, Bushell, S, Bradley, A, Tallant, C, Kiyani, W, Froese, D.S, Carpenter, E.S, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-01-29 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
5TVL
 
 | | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A | | Descriptor: | CHLORIDE ION, Foldase protein PrsA, GLYCEROL, ... | | Authors: | Borek, D, Yim, V, Kudritska, M, Wawrzak, Z, Stogios, P.J, Otwinowski, Z, Savchenko, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A
To Be Published
|
|
1PGV
 
 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
6AIB
 
 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5WPB
 
 | | Crystal structure of fragment 3-(3-(pyridin-2-ylmethoxy)quinoxalin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-{3-[(pyridin-2-yl)methoxy]quinoxalin-2-yl}propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Tempel, W, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-04 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
3RN4
 
 | | Crystal structure of iron-substituted Sod2 from Saccharomyces cerevisiae | | Descriptor: | FE (III) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Kang, Y, He, Y.-X, Cheng, W, Zhou, C.-Z, Li, W.-F. | | Deposit date: | 2011-04-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structures of native and Fe-substituted SOD2 from Saccharomyces cerevisiae
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6BPH
 
 | | Crystal structure of the chromodomain of RBBP1 | | Descriptor: | AT-rich interactive domain-containing protein 4A, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of chromo barrel domain of RBBP1.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
1LIH
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Scott, W, Yeh, J.I, Prive, G.G, Milburn, M. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
4PVJ
 
 | | Phl p 4 I153V variant, a glucose oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Pollen allergen Phl p 4.0202, ... | | Authors: | Zafred, D, Teufelberger, A, Keller, W, Macheroux, P. | | Deposit date: | 2014-03-17 | | Release date: | 2014-04-02 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
4C3F
 
 | |
