6UG0
 
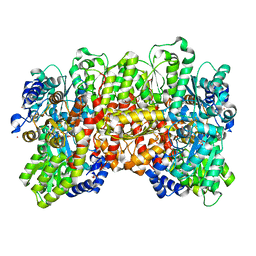 | | N2-bound Nitrogenase MoFe-protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Hu, Y, Ribbe, M.W. | | Deposit date: | 2019-09-25 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
7AD8
 
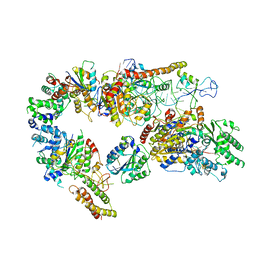 | | Core TFIIH-XPA-DNA complex with modelled p62 subunit | | Descriptor: | DNA (49-MER), DNA repair protein complementing XP-A cells, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2020-09-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The TFIIH subunits p44/p62 act as a damage sensor during nucleotide excision repair.
Nucleic Acids Res., 48, 2020
|
|
6UK5
 
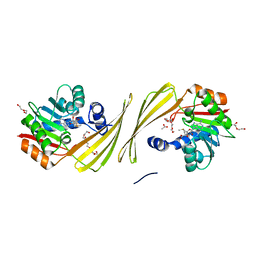 | | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis | | Descriptor: | ACETATE ION, CalS10, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alvarado, S.K, Miller, M.D, Xu, W, Wang, Z, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis
To Be Published
|
|
6U60
 
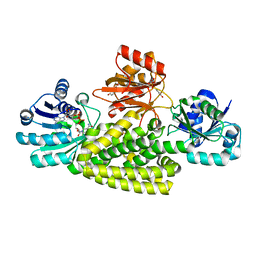 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD and L-tyrosine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Prephenate dehydrogenase, ... | | Authors: | Shabalin, I.G, Hou, J, Kutner, J, Grimshaw, S, Christendat, D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
6U6J
 
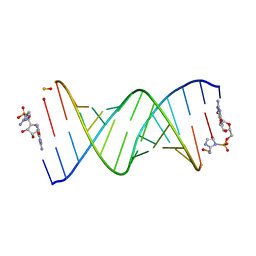 | | RNA-monomer complex containing pyrophosphate linkage | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(hydroxy)phosphoryl]guanosine, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(DPG))-3') | | Authors: | Zhang, W, Szostak, J.W, Giurgiu, C, Wright, T. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prebiotically Plausible "Patching" of RNA Backbone Cleavage through a 3'-5' Pyrophosphate Linkage.
J.Am.Chem.Soc., 141, 2019
|
|
6U8F
 
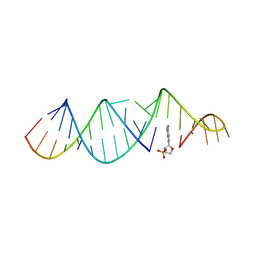 | | RNA hairpin, bound with TNA monomer | | Descriptor: | 2-azanyl-9-[(2~{R},3~{R},4~{S})-3-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-oxolan-2-yl]-1~{H}-purin-6-one, RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*CP*AP*UP*GP*AP*AP*AP*GP*U)-3'), RNA (5'-R(P*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6U5G
 
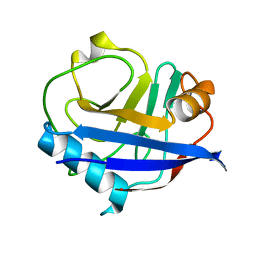 | | MicroED structure of a FIB-milled CypA Crystal | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Wolff, A.M, Martynowycz, M.W, Zhao, W, Gonen, T, Fraser, J.S, Thompson, M.C. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Iucrj, 7, 2020
|
|
6U7Y
 
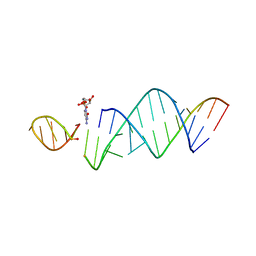 | | RNA hairpin structure containing one TNA nucleotide as template | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, RNA (5'-R(*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3'), RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*(TG)P*CP*CP*GP*AP*AP*AP*GP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6UE5
 
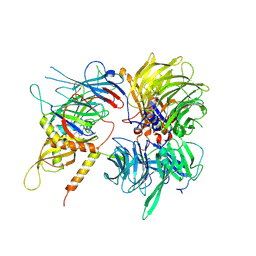 | | Crystal structure of full-length human DCAF15-DDB1-deltaPBP-DDA1-RBM39 in complex with 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzenesulfonamide | | Descriptor: | 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Knapp, M.S, Shu, W, Xie, L, Bussiere, D.E. | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
6TUM
 
 | |
6UKT
 
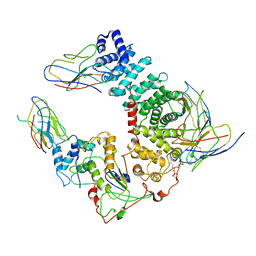 | | Cryo-EM structure of mammalian Ric-8A:Galpha(i):nanobody complex | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, NB8109, NB8117, ... | | Authors: | Mou, T.C, Zhang, K, Johnston, J.D, Chiu, W, Sprang, S.R. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to G alpha i1.
Nat Commun, 11, 2020
|
|
7AC9
 
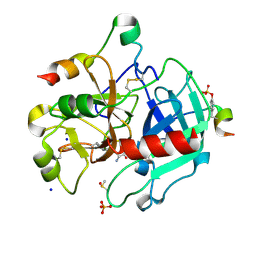 | | Thrombin in complex with D-arginine (j77) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-ARGININE, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Thrombin in complex with D-arginine (j77)
To be published
|
|
6U9I
 
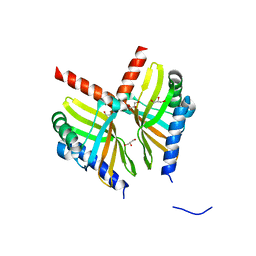 | | Crystal structure of BvnE pinacolase from Penicillium brevicompactum | | Descriptor: | BvnE, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Ye, Y, Du, L, Zhang, X, Newmister, S.A, McCauley, M, Alegre-Requena, J.V, Zhang, W, Mu, S, Minami, A, Fraley, A.E, Adrover-Castellano, M.L, Carney, N, Shende, V.V, Oikawa, H, Kato, H, Tsukamoto, S, Paton, R.S, Williams, R.M, Sherman, D.H, Li, S. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | Fungal-derived brevianamide assembly by a stereoselective semipinacolase.
Nat Catal, 3, 2020
|
|
6UBL
 
 | | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina | | Descriptor: | DynF, PALMITIC ACID | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Bhardwaj, M, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6UGH
 
 | |
6ZTG
 
 | | Spor protein DedD | | Descriptor: | Cell division protein DedD | | Authors: | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
6UI6
 
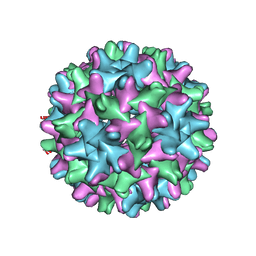 | | HBV T=3 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
7AHX
 
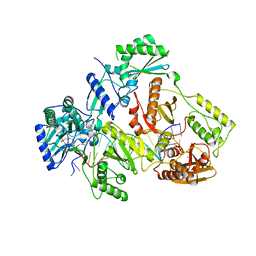 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND D-ASPARTATE TENOFOVIR WITH BOUND MANGANESE | | Descriptor: | D-Aspartate Tenofovir, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
6U5A
 
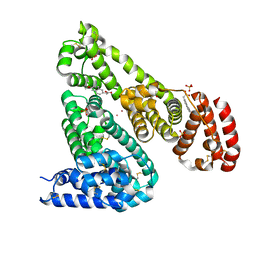 | | Crystal structure of Equine Serum Albumin complex with 6-MNA | | Descriptor: | (6-methoxynaphthalen-2-yl)acetic acid, SULFATE ION, Serum albumin, ... | | Authors: | Czub, M.P, Handing, K.B, Venkataramany, B.S, Cymborowski, M.T, Shabalin, I.G, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-27 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 63, 2020
|
|
6U5Z
 
 | |
7AQK
 
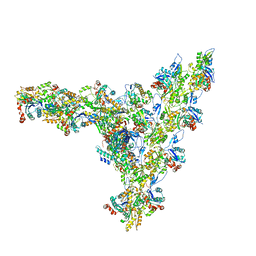 | | Model of the actin filament Arp2/3 complex branch junction in cells | | Descriptor: | Actin, alpha skeletal muscle, ACTA1, ... | | Authors: | Faessler, F, Dimchev, G, Hodirnau, V.V, Wan, W, Schur, F.K.M. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction.
Nat Commun, 11, 2020
|
|
7AII
 
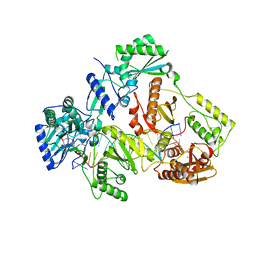 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-METHIONINE TENOFOVIR WITH BOUND MANGANESE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7B1A
 
 | | Myosin-II-AA mutant motor domain | | Descriptor: | 1,2-ETHANEDIOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Ewert, W, Preller, M. | | Deposit date: | 2020-11-24 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unraveling a Force-Generating Allosteric Pathway of Actomyosin Communication Associated with ADP and P i Release.
Int J Mol Sci, 22, 2020
|
|
6UJE
 
 | | Crystal structure of the Clostridial cellulose synthase subunit Z (CcsZ) from Clostridioides difficile | | Descriptor: | CALCIUM ION, Endoglucanase | | Authors: | Scott, W, Lowrance, B, Anderson, A.C, Weadge, J.T. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of the Clostridial cellulose synthase and characterization of the cognate glycosyl hydrolase, CcsZ.
Plos One, 15, 2020
|
|
6UR2
 
 | |
