8TA2
 
 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42 | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
4D08
 
 | | PDE2a catalytic domain in complex with a brain penetrant inhibitor | | Descriptor: | 1-(5-butoxypyridin-3-yl)-4-methyl-8-(morpholin-4-ylmethyl)[1,2,4]triazolo[4,3-a]quinoxaline, CGMP-DEPENDENT 3', 5'-CYCLIC PHOSPHODIESTERASE, ... | | Authors: | Buijnsters, P, Andres, J.I, DeAngelis, M, Langlois, X, Rombouts, F, Sanderson, W, Tresadern, G, Trabanco, A, VanHoof, G, VanRoosbroeck, Y. | | Deposit date: | 2014-04-24 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of a Potent, Selective, and Brain Penetrating Pde2 Inhibitor with Demonstrated Target Engagement.
Acs Med.Chem.Lett., 5, 2014
|
|
8TA3
 
 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
4GI7
 
 | | Crystal structure of Klebsiella pneumoniae pantothenate kinase in complex with a pantothenate analogue | | Descriptor: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-{3-oxo-3-[(pyridin-3-ylmethyl)amino]propyl}butanamide, ADENOSINE-5'-DIPHOSPHATE, Pantothenate kinase, ... | | Authors: | Li, B, Tempel, W, Smil, D, Bolshan, Y, Hong, B.S, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-08 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Klebsiella pneumoniae pantothenate kinase in complex with N-substituted pantothenamides.
Proteins, 81, 2013
|
|
4B4L
 
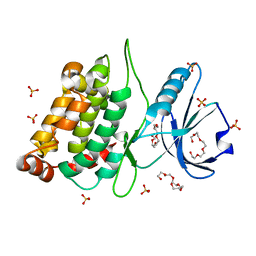 | | CRYSTAL STRUCTURE OF AN ARD DAP-KINASE 1 MUTANT | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, PENTAETHYLENE GLYCOL, SULFATE ION | | Authors: | Temmerman, K, Pogenberg, V, Jonko, W, Wilmanns, M. | | Deposit date: | 2012-07-31 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Pef/Y Substrate Recognition and Signature Motif Plays a Critical Role in Dapk-Related Kinase Activity.
Chem.Biol., 21, 2014
|
|
8TA6
 
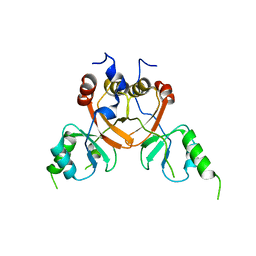 | | Cryo-EM structure of the human CLC-2 chloride channel C-terminal domain | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
4B50
 
 | |
4B3P
 
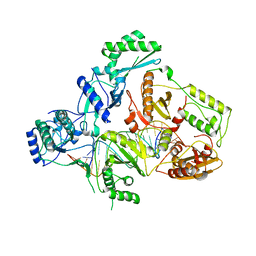 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | DNA, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.839 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4AXW
 
 | | CRYSTAL STRUCTURE OF MOUSE CADHERIN-23 EC1-2 AND PROTOCADHERIN-15 EC1- 2, FORM I 2.2A. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CADHERIN-23, CALCIUM ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of a Force-Conveying Cadherin Bond Essential for Inner-Ear Mechanotransduction
Nature, 492, 2012
|
|
8TES
 
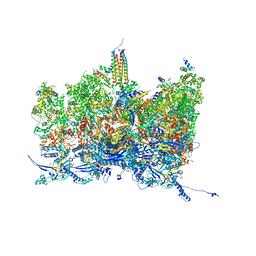 | | Human cytomegalovirus portal vertex, virion configuration 2 (VC2) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TET
 
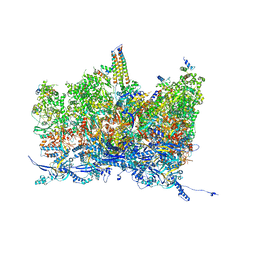 | | Human cytomegalovirus portal vertex, non-infectious enveloped particle (NIEP) configuration 1 (NC1) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TEW
 
 | | Human cytomegalovirus penton vertex, CVSC-bound configuration | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TOO
 
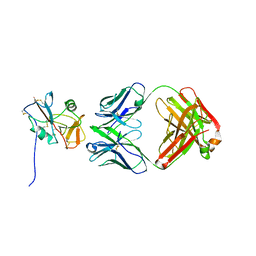 | | Crystal structure of Epstein-Barr virus gp42 in complex with antibody 4C12 | | Descriptor: | 4C12 heavy chain, 4C12 light chain, Glycoprotein 42 | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TNN
 
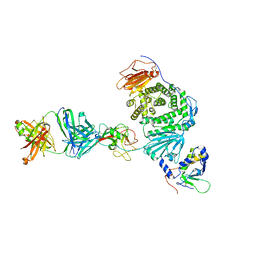 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with gp42 antibody A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A10 heavy chain, A10 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
4G2N
 
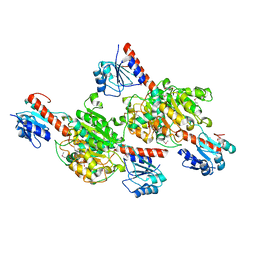 | | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66 | | Descriptor: | CHLORIDE ION, D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66
To be Published
|
|
4G9Z
 
 | | Lassa nucleoprotein with dsRNA reveals novel mechanism for immune suppression | | Descriptor: | Nucleoprotein, RNA (5'-R(P*GP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2012-07-24 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
4B3O
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, 5'-D(*CP*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP *TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3', 5'-R(*AP*UP*GP*AP*3DRP*GP*GP*CP*CP*AP*CP*AP*AP*UP*AP *AP*CP*UP*AP*UP*AP*GP*GP*CP*AP*UP*A)-3', ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
8TEP
 
 | | Human cytomegalovirus portal vertex, virion configuration 1 (VC1) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Inner tegument protein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
4G73
 
 | | Crystal structure of NDH with NADH and Quinone | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
4GES
 
 | | crystal structure of GFP-TYR151PYZ with an unnatural amino acid incorporation | | Descriptor: | Green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4GF1
 
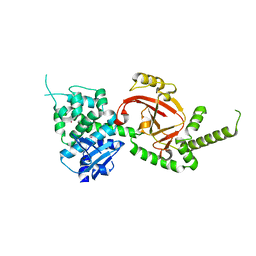 | | Crystal Structure of Certhrax | | Descriptor: | Putative ADP-ribosyltransferase Certhrax, UNKNOWN ATOM OR ION | | Authors: | Hong, B.S, Dimov, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
8TEU
 
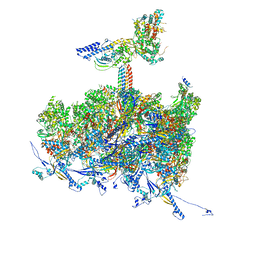 | | Human cytomegalovirus portal vertex, non-infectious enveloped particle (NIEP) configuration 2 - inverted (NC2-inv) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TNT
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with antibodies F-2-1 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 heavy chain, 769C2 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
4B27
 
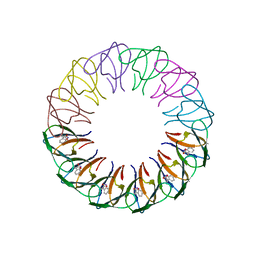 | | Trp RNA-binding attenuation protein: modifying symmetry and stability of a circular oligomer | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Bayfield, O.W, Chen, C, Patterson, A.R, Luan, W, Smits, C, Gollnick, P, Antson, A.A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Trp RNA-Binding Attenuation Protein: Modifying Symmetry and Stability of a Circular Oligomer.
Plos One, 7, 2012
|
|
4B5X
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase (HpaI), mutant D42A | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
