8QVU
 
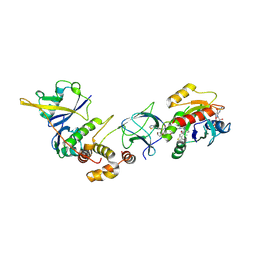 | | Crystal Structure of ligand ACBI3 in complex with KRAS G12D C118S GDP and pVHL:ElonginC:ElonginB complex | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Wijaya, A.J, Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Targeting cancer with small molecule pan-KRAS degraders
Biorxiv, 2023
|
|
1QFF
 
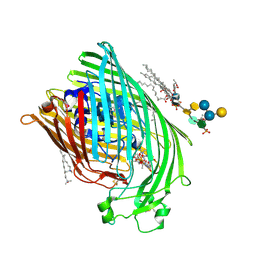 | | E. COLI FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) IN COMPLEX WITH BOUND FERRICHROME-IRON | | Descriptor: | 2-AMINO-VINYL-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Ferguson, A.D, Hofmann, E, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
8QW6
 
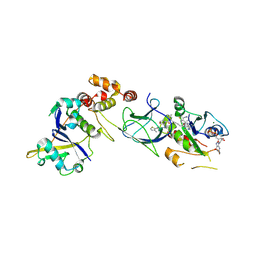 | | Crystal Structure of compound 3 in complex with KRAS G12V C118S GDP and pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-1-[(2S)-2-[6-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]hexanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting cancer with small molecule pan-KRAS degraders
Biorxiv, 2023
|
|
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QPR
 
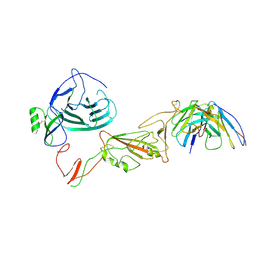 | | SARS-CoV-2 S protein bound to human neutralising antibody UZGENT_G5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG heavy chain - FAB, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
6NFT
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (4-oxoquinazolin-3(4H)-yl)acetic acid | | Descriptor: | (4-oxoquinazolin-3(4H)-yl)acetic acid, 1,2-ETHANEDIOL, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Mann, M.K, Tempel, W, Bountra, C, Arrowmsmith, C.M, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
6NFX
 
 | | MBTD1 MBT repeats | | Descriptor: | GLYCEROL, MBT domain-containing protein 1,Enhancer of polycomb homolog 1, SODIUM ION, ... | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-21 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for EPC1-Mediated Recruitment of MBTD1 into the NuA4/TIP60 Acetyltransferase Complex.
Cell Rep, 30, 2020
|
|
8QQ0
 
 | | SARS-CoV-2 S protein bound to neutralising antibody UZGENT_A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
6NYG
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2a (OA-2a) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ZEA
 
 | | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{R})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, (1~{S})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, 1,2-ETHANEDIOL, ... | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline
To Be Published
|
|
6NCH
 
 | | Crystal structure of CDP-Chase: Raster data collection | | Descriptor: | D-ribose, PHOSPHATE ION, Phosphohydrolase (MutT/nudix family protein), ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6Z7U
 
 | | Myosin-II motor domain complexed with blebbistatin in a new ADP-release conformation | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ewert, W, Preller, M. | | Deposit date: | 2020-06-01 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and Computational Insights into a Blebbistatin-Bound Myosin•ADP Complex with Characteristics of an ADP-Release Conformation along the Two-Step Myosin Power Stoke.
Int J Mol Sci, 21, 2020
|
|
6NCO
 
 | | Fragment-based Discovery of an apoE4 Stabilizer | | Descriptor: | 1-[5-chloro-4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]cyclobutane-1-carboximidamide, Apolipoprotein E | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer.
J.Med.Chem., 62, 2019
|
|
6NGG
 
 | |
6NRA
 
 | | hTRiC-hPFD Class1 (No PFD) | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6ZCT
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
6ZDR
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with Chromone 4d | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Verdon, G, Jespers, W, Azuaje, J, Majellaro, M, Keranen, H, Garcia-mera, X, Congreve, M, Deflorian, F, de Graaf, C, Zhukov, A, Dore, A, Mason, J, Aqvist, J, Cooke, R, Sotelo, E, Gutierrez-de-Teran, H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | X-Ray Crystallography and Free Energy Calculations Reveal the Binding Mechanism of A 2A Adenosine Receptor Antagonists.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6NJE
 
 | | Crystal structure of the motor domain of human kinesin family member 22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Kinesin-like protein KIF22, ... | | Authors: | Walker, B.C, Zhu, H, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Park, H, Cochran, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-03 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the motor domain of human kinesin family member 22
To Be Published
|
|
1QNJ
 
 | | THE STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT ATOMIC RESOLUTION (1.1 A) | | Descriptor: | ELASTASE, SODIUM ION, SULFATE ION | | Authors: | Wurtele, M, Hahn, M, Hilpert, K, Hohne, W. | | Deposit date: | 1999-10-15 | | Release date: | 2000-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structure of Native Porcine Pancreatic Elastase at 1.1 A
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6O0B
 
 | | Structural and Mechanistic Insights into CO2 Activation by Nitrogenase Iron Protein | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein | | Authors: | Rettberg, L.A, Stiebritz, M.T, Kang, W, Lee, C.C, Ribbe, M.W, Hu, Y. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Insights into CO2Activation by Nitrogenase Iron Protein.
Chemistry, 25, 2019
|
|
6O0W
 
 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
8RM7
 
 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: MMTV-177 GRE/ARE | | Descriptor: | Isoform 2 of Androgen receptor, MMTV-177 GRE/ARE Chain C, MMTV-177 GRE/ARE, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
8RM6
 
 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: C3(1)ARE | | Descriptor: | C3(1)ARE_Chain C, C3(1)ARE_Chain D, Isoform 2 of Androgen receptor, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
