4PMQ
 
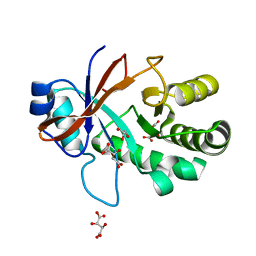 | | Crystal structure of the Mycobacterium tuberculosis Tat-secreted protein Rv2525c in complex with L-tartrate (orthorhombic crystal form) | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Tat-secreted protein Rv2525c | | Authors: | Bellinzoni, M, Haouz, A, Shepard, W, Alzari, P.M. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural studies suggest a peptidoglycan hydrolase function for the Mycobacterium tuberculosis Tat-secreted protein Rv2525c.
J.Struct.Biol., 188, 2014
|
|
9B6K
 
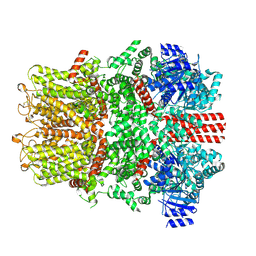 | | Cryo-EM structure of the mouse TRPM8 channel in complex with Ca2+ in the absence of PI(4,5)P2 | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
3VPV
 
 | |
4PNS
 
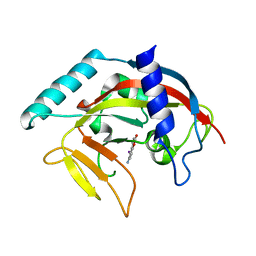 | | Crystal Structure of human Tankyrase 2 in complex with INH2BP. | | Descriptor: | 6-amino-5-iodo-2H-chromen-2-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4EA3
 
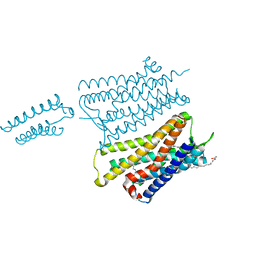 | | Structure of the N/OFQ Opioid Receptor in Complex with a Peptide Mimetic | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-benzyl-N-[3-(1'H,3H-spiro[2-benzofuran-1,4'-piperidin]-1'-yl)propyl]-D-prolinamide, ... | | Authors: | Thompson, A.A, Liu, W, Chun, E, Katritch, V, Wu, H, Vardy, E, Huang, X.P, Trapella, C, Guerrini, R, Calo, G, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic.
Nature, 485, 2012
|
|
4P7A
 
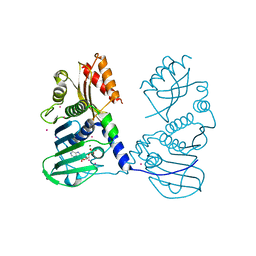 | | Crystal Structure of human MLH1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Mlh1, MAGNESIUM ION, ... | | Authors: | Tempel, W, Lam, R, Zeng, H, Walker, J.R, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human MLH1 N-terminus: implications for predisposition to Lynch syndrome.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4P8W
 
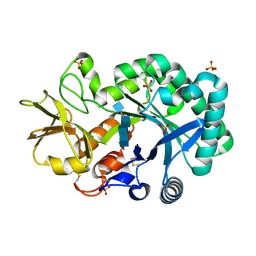 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc4) were solved to resolutions of 1.9 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4ECW
 
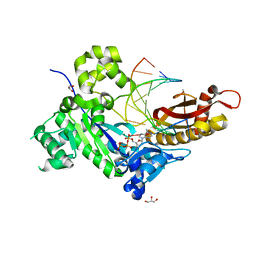 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 250 sec | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4EFE
 
 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
4EGX
 
 | | Crystal structure of KIF1A CC1-FHA tandem | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Kinesin-like protein KIF1A | | Authors: | Yu, J, Huo, L, Yue, Y, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-02 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
4E4G
 
 | | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
4E6M
 
 | | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium)
To be Published
|
|
4PDV
 
 | | Structure of K+ selective NaK mutant in barium and potassium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BARIUM ION, POTASSIUM ION, ... | | Authors: | Lam, Y, Zeng, W, Sauer, D.B, Jiang, Y. | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | High Resolution Structural Views of Rubidium, Cesium and Barium Binding within a Potassium Selective Channel Filter
To Be Published
|
|
4ETA
 
 | | Lysozyme, room temperature, 400 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ECQ
 
 | | Human DNA polymerase eta- DNA ternary complex: AT crystal at pH6.8(K+ MES) with 1 Ca2+ ion | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*TP*A)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ED1
 
 | | Human DNA polymerase eta - DNA ternary complex: AT crystal at pH 7.0 (Na+ MES) with 1 Ca2+ ion | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
3T7V
 
 | | Crystal structure of methylornithine synthase (PylB) | | Descriptor: | 5-amino-D-isoleucine, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Quitterer, F, List, A, Eisenreich, W, Bacher, A, Groll, M. | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4EIY
 
 | | Crystal structure of the chimeric protein of A2aAR-BRIL in complex with ZM241385 at 1.8A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Liu, W, Chun, E, Thompson, A.A, Chubukov, P, Xu, F, Katritch, V, Han, G.W, Heitman, L.H, Ijzerman, A.P, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-04-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for allosteric regulation of GPCRs by sodium ions.
Science, 337, 2012
|
|
3SZP
 
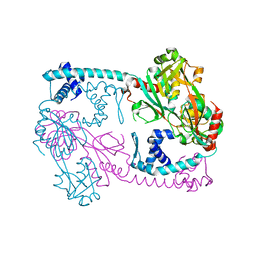 | | Full-length structure of the Vibrio cholerae virulence activator, AphB, a member of the LTTR protein family | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Taylor, J.L, De Silva, R.S, Kovacikova, G, Lin, W, Taylor, R.K, Skorupski, K, Kull, F.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH.
Mol.Microbiol., 83, 2012
|
|
3T0G
 
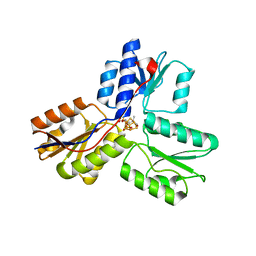 | | IspH:HMBPP (substrate) structure of the T167C mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
4EJQ
 
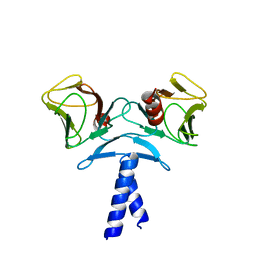 | | Crystal structure of KIF1A C-CC1-FHA | | Descriptor: | Kinesin-like protein KIF1A | | Authors: | Huo, L, Yue, Y, Ren, J, Yu, J, Liu, J, Yu, Y, Ye, F, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-06 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
3T1C
 
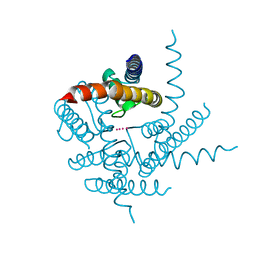 | | Crystal Structure of NaK Channel D66Y Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4E8C
 
 | | Crystal structure of streptococcal beta-galactosidase in complex with galactose | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35, ... | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
4NMH
 
 | | 11-beta-HSD1 in complex with a 3,3-Di-methyl-azetidin-2-one | | Descriptor: | (4S)-4-(2-methoxyphenyl)-3,3-dimethyl-1-[3-(methylsulfonyl)phenyl]azetidin-2-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McCoull, W, Augustin, M, Blake, C, Ertan, A, Kilgour, E.K, Krapp, S, Moore, J.E, Newcombe, N.J, Packer, M.J, Rees, A, Revill, J, Scott, J.S, Selmi, N, Gerhardt, S, Ogg, D.J, Steinbacher, S, Whittamore, P.R.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification and optimisation of 3,3-dimethyl-azetidin-2-ones as potent and selective inhibitors of 11 beta-hydroxysteroid dehydrogenase type 1 (11-beta-HSD1)
TO BE PUBLISHED
|
|
4EB3
 
 | | Crystal structure of IspH in complex with iso-HMBPP | | Descriptor: | 3-(hydroxymethyl)but-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Wang, W, Wang, K, Span, I, Bacher, A, Groll, M, Oldfield, E. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Are free radicals involved in IspH catalysis? An EPR and crystallographic investigation.
J.Am.Chem.Soc., 134, 2012
|
|
