1BTH
 
 | |
2BQP
 
 | | THE STRUCTURE OF THE PEA LECTIN-D-GLUCOPYRANOSE COMPLEX | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEA LECTIN), ... | | Authors: | Ruzeinikov, S.N, Mikhailova Yu, I, Tsygannik, I.N, Pangborn, W, Duax, W, Pletnev, V.Z. | | Deposit date: | 1998-12-08 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Pea Lectin-D-Glucopyranose Complex at a 1.9 A Resolution
RUSS.J.BIOORGANIC CHEM., 23, 1997
|
|
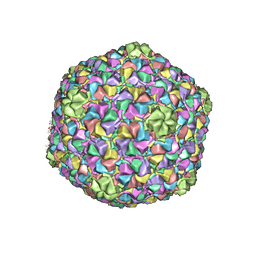
3J40
 
 | | Validated Near-Atomic Resolution Structure of Bacteriophage Epsilon15 Derived from Cryo-EM and Modeling | | Descriptor: | gp10, gp7 | | Authors: | Baker, M.L, Hryc, C.F, Zhang, Q, Wu, W, Jakana, J, Haase-Pettingell, C, Afonine, P.V, Adams, P.D, King, J.A, Jiang, W, Chiu, W. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
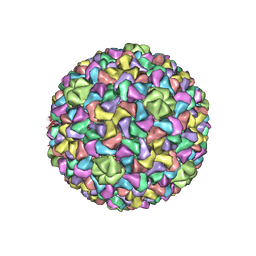
3J7W
 
 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8D56
 
 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D55
 
 | | Closed state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8GRA
 
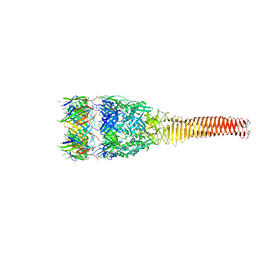 | |
3J7X
 
 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8D5A
 
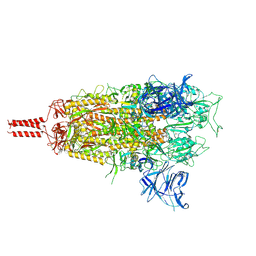 | | Middle state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3J7V
 
 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1CXR
 
 | |
2VKE
 
 | |
6KNM
 
 | | Apelin receptor in complex with single domain antibody | | Descriptor: | Apelin receptor,Rubredoxin,Apelin receptor, Single domain antibody JN241, ZINC ION | | Authors: | Ma, Y.B, Ding, Y, Song, X, Ma, X, Li, X, Zhang, N, Song, Y, Sun, Y, Shen, Y, Zhong, W, Hu, L.A, Ma, Y.L, Zhang, M.Y. | | Deposit date: | 2019-08-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-guided discovery of a single-domain antibody agonist against human apelin receptor.
Sci Adv, 6, 2020
|
|
6K0V
 
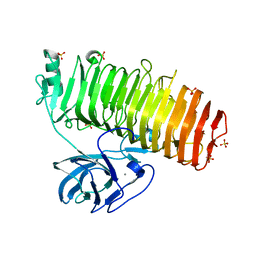 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K57
 
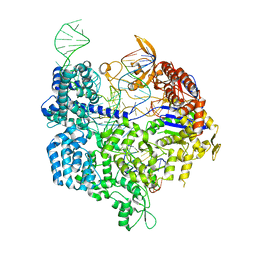 | | Crystal structure of dCas9 in complex with sgRNA and DNA (CGA PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9, non-target DNA, sgRNA, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-28 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6KEA
 
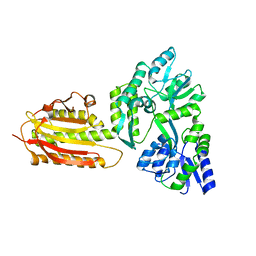 | | crystal structure of MBP-tagged REV7-IpaB complex | | Descriptor: | Maltose-binding periplasmic protein,LINKER,hREV7,LINKER,Invasin IpaB,hREV3 | | Authors: | Wang, X, Pernicone, N, Pertz, L, Hua, D.P, Zhang, T.Q, Listovsky, T, Xie, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | REV7 has a dynamic adaptor region to accommodate small GTPase RAN/ShigellaIpaB ligands, and its activity is regulated by the RanGTP/GDP switch.
J.Biol.Chem., 294, 2019
|
|
2YHY
 
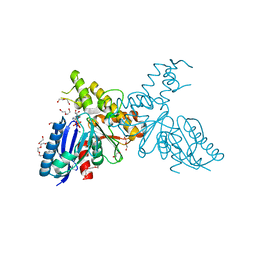 | | Structure of N-Acetylmannosamine kinase in complex with N- acetylmannosamine and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Reutter, W, Fan, H, Saenger, W, Moniot, S. | | Deposit date: | 2011-05-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
6KJJ
 
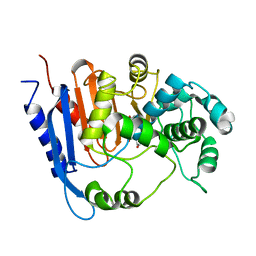 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | 4-(2-acetamidoethylsulfanyl)-4-oxidanylidene-butanoic acid, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6DVR
 
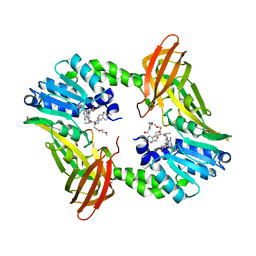 | | Crystal structure of human CARM1 with (R)-SKI-72 | | Descriptor: | (2R,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-methoxyphenyl)ethyl]hexanamide (non-preferred name), 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Zeng, H, Hutchinson, A, Seitova, A, Luo, M, Cai, X.C, Ke, W, Wang, J, Shi, C, Zheng, W, Lee, J.P, Ibanez, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of human CARM1 with (R)-SKI-72
to be published
|
|
6K2L
 
 | | Crystal structure of the Siderophore-interacting protein SipS from Aeromonas hydrophila | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Siderophore-interacting protein | | Authors: | Shang, F, Lan, J, Liu, W, Xu, Y. | | Deposit date: | 2019-05-14 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Siderophore-interacting protein SIP from Aeromonas hydrophila.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
6NU4
 
 | | Solution structure of the Arabidopsis thaliana RALF8 peptide | | Descriptor: | Protein RALF-like 8 | | Authors: | Lee, W, Markley, J.L, Frederick, R.O, Miyoshi, H, Tonelli, M, Cornilescu, G, Cornilescu, C, Sussman, M.R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Function and solution structure of the Arabidopsis thaliana RALF8 peptide.
Protein Sci., 28, 2019
|
|
6K0S
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6KFI
 
 | | NMR solution structure of the 1:1 complex of Tel26 G-quadruplex and a tripodal cationic fluorescent probe NBTE | | Descriptor: | 4,4',4''-(nitrilotris(benzene-4,1-diyl))tris(1-ethylpyridin-1-ium) iodide, G-quadruplex DNA (26-MER) | | Authors: | Liu, W, Liu, L.Y, Mao, Z.W. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6K4Q
 
 | | Crystal structure of xCas9 in complex with sgRNA and DNA (CGG PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*CP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-26 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6KFJ
 
 | | NMR solution structure of the 1:1 complex of wtTel26 G-quadruplex and a tripodal cationic fluorescent probe NBTE | | Descriptor: | 4,4',4''-(nitrilotris(benzene-4,1-diyl))tris(1-ethylpyridin-1-ium) iodide, G-quadruplex DNA wtTel26 | | Authors: | Liu, W, Liu, L.Y, Mao, Z.W. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
