5ZE2
 
 | | Hairpin Complex, RAG1/2-hairpin 12RSS/23RSS complex in 5mM Mn2+ for 2 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (31-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
2O2S
 
 | | The structure of T. gondii enoyl acyl carrier protein reductase in complex with NAD and triclosan | | Descriptor: | Enoyl-acyl carrier reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Muench, S.P, Prigge, S.T, McLeod, R, Rafferty, J.B, Kirisits, M.J, Roberts, C.W, Mui, E.J, Rice, D.W. | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Studies of Toxoplasma gondii and Plasmodium falciparum enoyl acyl carrier protein reductase and implications for the development of antiparasitic agents
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
3BAE
 
 | | Crystal structure of Fab WO2 bound to the N terminal domain of Amyloid beta peptide (1-28) | | Descriptor: | Amyloid Beta Peptide, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Parker, M.W. | | Deposit date: | 2007-11-07 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
8PX2
 
 | |
6T3J
 
 | | Dual Epitope Targeting by Anti-DR5 Antibodies | | Descriptor: | IgG1-hDR5-01-Heavy Chain, IgG1-hDR5-01-Light Chain, IgG1-hDR5-05-Heavy Chain, ... | | Authors: | Tauchert, M.J, Augustin, M, Krapp, S, Overdijk, M.B, Breij, E.C.W, Hibbert, R.G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Dual Epitope Targeting and Enhanced Hexamerization by DR5 Antibodies as a Novel Approach to Induce Potent Antitumor Activity Through DR5 Agonism.
Mol.Cancer Ther., 19, 2020
|
|
8PX8
 
 | |
5YYL
 
 | | Structure of Major Royal Jelly Protein 1 Oligomer | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tian, W, Chen, Z. | | Deposit date: | 2017-12-10 | | Release date: | 2018-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architecture of the native major royal jelly protein 1 oligomer.
Nat Commun, 9, 2018
|
|
3B50
 
 | | Structure of H. influenzae sialic acid binding protein bound to Neu5Ac. | | Descriptor: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein siaP | | Authors: | Ramaswamy, S, Coussens, N.P, Apicella, M.A, Johnston, J.W. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of the N-Acetyl-5-neuraminic Acid-binding Site of the Extracytoplasmic Solute Receptor (SiaP) of Nontypeable Haemophilus influenzae Strain 2019
J.Biol.Chem., 283, 2008
|
|
1XYS
 
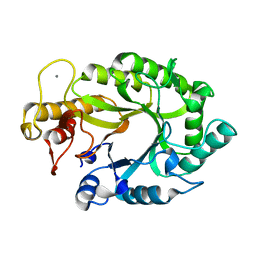 | | CATALYTIC CORE OF XYLANASE A E246C MUTANT | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1994-09-02 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the catalytic core of the family F xylanase from Pseudomonas fluorescens and identification of the xylopentaose-binding sites.
Structure, 2, 1994
|
|
5M0M
 
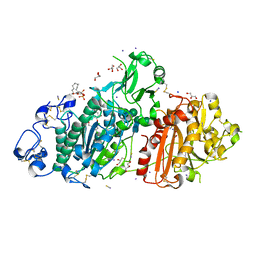 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
5M0S
 
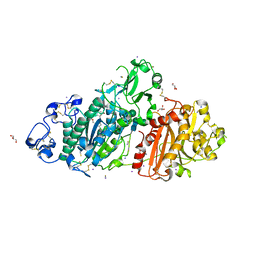 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, GLYCEROL, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
7V3A
 
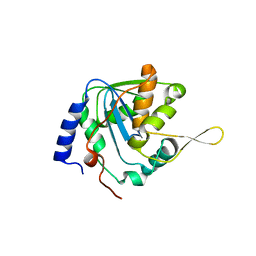 | | Crystal structure of apo-NP exonuclease C409A | | Descriptor: | MAGNESIUM ION, Nucleoprotein, ZINC ION | | Authors: | Hsiao, Y.Y, Huang, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites.
Jacs Au, 1, 2021
|
|
7V38
 
 | | Crystal structure of NP exonuclease-PCMPS complex | | Descriptor: | Nucleoprotein, PARA-MERCURY-BENZENESULFONIC ACID, ZINC ION | | Authors: | Hsiao, Y.Y, Huang, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites.
Jacs Au, 1, 2021
|
|
7V3C
 
 | | Crystal structure of NP exonuclease C409A-PCMB complex | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, MAGNESIUM ION, Nucleoprotein, ... | | Authors: | Hsiao, Y.Y, Huang, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites.
Jacs Au, 1, 2021
|
|
7V39
 
 | | Crystal structure of NP exonuclease-PCMB complex | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Nucleoprotein, ZINC ION | | Authors: | Hsiao, Y.Y, Huang, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites.
Jacs Au, 1, 2021
|
|
7V3B
 
 | | Crystal structure of NP exonuclease C409A-PCMPS complex | | Descriptor: | MAGNESIUM ION, Nucleoprotein, PARA-MERCURY-BENZENESULFONIC ACID, ... | | Authors: | Hsiao, Y.Y, Huang, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites.
Jacs Au, 1, 2021
|
|
6MF0
 
 | | Crystal Structure Determination of Human/Porcine Chimera Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19999886 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
6RNW
 
 | |
5O4L
 
 | | Crystal structure of P450 CYP121 in complex with compound 6a. | | Descriptor: | 1-[(4-fluorophenyl)methyl]-4-(3-imidazol-1-ylpropyl)piperazin-2-one, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-05-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
5O65
 
 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5O6D
 
 | | Structure of ScPif1 in complex with polydT and ATPgS | | Descriptor: | ATP-dependent DNA helicase PIF1, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
5AGW
 
 | | Bcl-2 alpha beta-1 complex | | Descriptor: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11 | | Authors: | Smith, B.J, F Lee, E, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Alpha Beta Peptide Foldamers Targeting Intracellular Protein-Protein Interactions with Activity on Living Cells
J.Am.Chem.Soc., 137, 2015
|
|
2XA3
 
 | | crystal structure of the broadly neutralizing llama VHH D7 and its mode of HIV-1 gp120 interaction | | Descriptor: | LLAMA HEAVY CHAIN ANTIBODY D7, SULFATE ION | | Authors: | Hinz, A, Lutje Hulsik, D, Forsman, A, Koh, W, Belrhali, H, Gorlani, A, de Haard, H, Weiss, R.A, Verrips, T, Weissenhorn, W. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the neutralizing Llama V(HH) D7 and its mode of HIV-1 gp120 interaction.
PLoS ONE, 5, 2010
|
|
