2W62
 
 | | Saccharomyces cerevisiae Gas2p in complex with laminaripentaose | | Descriptor: | 1,4-BUTANEDIOL, GLYCOLIPID-ANCHORED SURFACE PROTEIN 2, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Schuettelkopf, A.W, Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Mechanisms of Yeast Cell Wall Glucan Remodeling.
J.Biol.Chem., 284, 2009
|
|
4AA6
 
 | | The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation | | Descriptor: | 5'-D(*CP*TP*AP*AP*GP*TP*CP*AP*CP*AP*GP*TP*GP*AP *CP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*GP*TP*CP*AP*CP*TP*GP*TP*GP*AP *CP*TP*TP*A)-3', ESTROGEN RECEPTOR, ... | | Authors: | Schwabe, J.W, Chapman, L, Rhodes, D. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation.
Structure, 3, 1995
|
|
7FAH
 
 | | Immune complex of head region of CA09 HA and neutralizing antibody 12H5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Li, T.T, Xue, W.H, Gu, Y, Li, S.W. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
4AB3
 
 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
8T1O
 
 | | AP2 bound to MSP2N2 nanodisc with Tgn38 cargo peptide; composite map | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Cannon, K.S, Reta, S. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lipid nanodiscs as a template for high-resolution cryo-EM structures of peripheral membrane proteins.
J.Struct.Biol., 215, 2023
|
|
7KYQ
 
 | |
2N8A
 
 | | 1H, 13C and 15N chemical shift assignments and solution structure for PARP-1 F1F2 domains in complex with a DNA single-strand break | | Descriptor: | DNA (45-MER), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Wu, W. | | Deposit date: | 2015-10-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1.
Mol.Cell, 60, 2015
|
|
8SW0
 
 | | Puromycin sensitive aminopeptidase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Puromycin-sensitive aminopeptidase, ... | | Authors: | Rodgers, D.W, Sampath, S. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of puromycin-sensitive aminopeptidase and polyglutamine binding.
Plos One, 18, 2023
|
|
7KYS
 
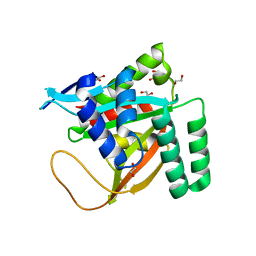 | | Crystal structure of human BCCIP beta (Native2) | | Descriptor: | 1,2-ETHANEDIOL, BRCA2 and CDKN1A-interacting protein, DI(HYDROXYETHYL)ETHER | | Authors: | Choi, W.S, Liu, B, Shen, Z, Yang, W. | | Deposit date: | 2020-12-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human BCCIP and implications for binding and modification of partner proteins.
Protein Sci., 30, 2021
|
|
7D0G
 
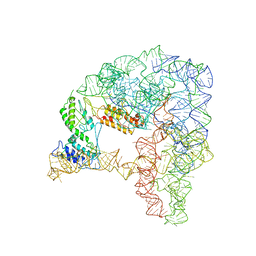 | | Cryo-EM structure of a pre-catalytic group II intron | | Descriptor: | Group II intron-encoded protein LtrA, RNA (714-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
7L27
 
 | |
2N59
 
 | | Solution Structure of R. palustris CsgH | | Descriptor: | Putative uncharacterized protein CsgH | | Authors: | Hawthorne, W.J, Taylor, J.D, Escalera-Maurer, A, Lambert, S, Koch, M, Scull, N, Sefer, L, Xu, Y, Matthews, S.J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-05-11 | | Method: | SOLUTION NMR | | Cite: | Electrostatically-guided inhibition of Curli amyloid nucleation by the CsgC-like family of chaperones.
Sci Rep, 6, 2016
|
|
6D69
 
 | |
8T9L
 
 | | Pom34-Pom152 membrane attachment site yeast NPC | | Descriptor: | Nucleoporin POM152, Nucleoporin POM34 | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-06-24 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
8GTX
 
 | |
3PTB
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | BENZAMIDINE, BETA-TRYPSIN, CALCIUM ION | | Authors: | Bode, W, Schwager, P, Walter, J. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
2MUW
 
 | | NOE-based model of the influenza A virus N31S mutant (19-49) bound to drug 11 | | Descriptor: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
4B6F
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (4-phenoxyphenyl)methylazanium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, ... | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
8TIE
 
 | | Double nuclear outer ring of Nup84-complexes from the yeast NPC | | Descriptor: | NUP133 isoform 1, NUP145 isoform 1, Nucleoporin NUP120, ... | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-07-19 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
8G0W
 
 | |
4Q3Q
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase, GLYCEROL, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
1LUL
 
 | | DB58, A LEGUME LECTIN FROM DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, LECTIN DB58, MANGANESE (II) ION | | Authors: | Hamelryck, T.W, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M, Loris, R. | | Deposit date: | 1998-06-30 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
1MPS
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH PHE M 197 REPLACED WITH ARG AND TYR M 177 REPLACED WITH PHE (CHAIN M, Y177F, F197R) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Mcauley-Hecht, K.E, Fyfe, P.K, Ridge, J.P, Prince, S, Hunter, C.N, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 1998-03-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural studies of wild-type and mutant reaction centers from an antenna-deficient strain of Rhodobacter sphaeroides: monitoring the optical properties of the complex from bacterial cell to crystal.
Biochemistry, 37, 1998
|
|
3QB4
 
 | |
4AX6
 
 | | HYPOCREA JECORINA CEL6A D221A MUTANT SOAKED WITH 6-CHLORO-4- PHENYLUMBELLIFERYL-BETA-CELLOBIOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloranyl-7-oxidanyl-4-phenyl-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-10 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
