3KTU
 
 | |
3UBT
 
 | |
3IH7
 
 | |
1HU0
 
 | | CRYSTAL STRUCTURE OF AN HOGG1-DNA BOROHYDRIDE TRAPPED INTERMEDIATE COMPLEX | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-OXOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2001-01-03 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Product-Assisted Catalysis in base-excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
6O8F
 
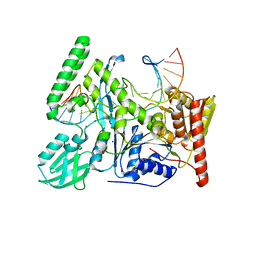 | | Crystal structure of UvrB bound to duplex DNA | | Descriptor: | ACETATE ION, CHLORIDE ION, DNA (5'-D(*GP*CP*CP*GP*TP*AP*TP*GP*CP*CP*AP*AP*TP*CP*TP*AP*GP*AP*GP*C)-3'), ... | | Authors: | Lee, S.-J, Verdine, G.L. | | Deposit date: | 2019-03-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Mechanism of DNA Lesion Homing and Recognition by the Uvr Nucleotide Excision Repair System.
Res, 2019, 2019
|
|
6OQA
 
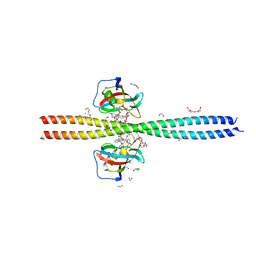 | | Crystal structure of CEP250 bound to FKBP12 in the presence of FK506-like novel natural product | | Descriptor: | (3R,4E,7E,10R,11S,12R,13S,16R,17R,24aS)-11,17-dihydroxy-10,12,16-trimethyl-3-[(2R)-1-phenylbutan-2-yl]-6,9,10,11,12,13,14,15,16,17,22,23,24,24a-tetradecahydro-3H-13,17-epoxypyrido[2,1-c][1,4]oxazacyclohenicosine-1,18,19(21H)-trione, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Lee, S.-J, Shigdel, U.K, Townson, S.A, Verdine, G.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1ZGW
 
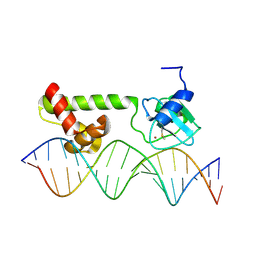 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
1ZXM
 
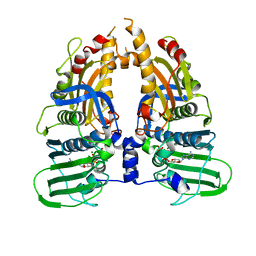 | | Human Topo IIa ATPase/AMP-PNP | | Descriptor: | DNA topoisomerase II, alpha isozyme, MAGNESIUM ION, ... | | Authors: | Wei, H, Ruthenburg, A.J, Bechis, S.K, Verdine, G.L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Nucleotide-dependent Domain Movement in the ATPase Domain of a Human Type IIA DNA Topoisomerase.
J.Biol.Chem., 280, 2005
|
|
4DJS
 
 | | Structure of beta-catenin in complex with a stapled peptide inhibitor | | Descriptor: | Catenin beta-1, stapled peptide RRWPQ(MK8)ILD(MK8)HVRRVWR | | Authors: | Bowman, B.R, Grossmann, T.N, Yeh, J.T.-H, Verdine, G.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.029 Å) | | Cite: | Inhibition of oncogenic Wnt signaling through direct targeting of beta-catenin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5KN9
 
 | | MutY N-terminal domain in complex with DNA containing an intrahelical oxoG:A base-pair | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Wang, L, Chakravarthy, S, Verdine, G.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
5KN8
 
 | | MutY N-terminal domain in complex with undamaged DNA | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Wang, L, Chakravarthy, S, Verdine, G.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
4YPR
 
 | | Crystal Structure of D144N MutY bound to its anti-substrate | | Descriptor: | A/G-specific adenine glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(*T*GP*TP*CP*CP*AP*CP*GP*TP*CP*T)-3'), ... | | Authors: | Wang, L, Lee, S, Verdine, G.L. | | Deposit date: | 2015-03-13 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase.
J.Biol.Chem., 290, 2015
|
|
4YOQ
 
 | | Crystal Structure of MutY bound to its anti-substrate | | Descriptor: | A/G-specific adenine glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(*T*GP*TP*CP*CP*AP*CP*GP*TP*CP*T)-3'), ... | | Authors: | Wang, L, Lee, S, Verdine, G.L. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase.
J.Biol.Chem., 290, 2015
|
|
4YPH
 
 | | Crystal Structure of MutY bound to its anti-substrate with the disulfide cross-linker reduced | | Descriptor: | A/G-specific adenine glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(TP*GP*TP*CP*CP*AP*CP*GP*TP*CP*T)-3'), ... | | Authors: | Wang, L, Lee, S, Verdine, G.L. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase.
J.Biol.Chem., 290, 2015
|
|
6O8E
 
 | | Crystal structure of UvrB bound to duplex DNA with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*GP*CP*CP*GP*TP*AP*TP*GP*CP*CP*AP*AP*TP*CP*TP*AP*GP*AP*GP*C)-3'), ... | | Authors: | Lee, S.-J, Verdine, G.L. | | Deposit date: | 2019-03-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Mechanism of DNA Lesion Homing and Recognition by the Uvr Nucleotide Excision Repair System.
Res, 2019, 2019
|
|
6O8H
 
 | | Crystal structure of UvrB mutant bound to duplex DNA | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*GP*CP*GP*CP*GP*AP*TP*GP*GP*AP*GP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*CP*GP*CP*GP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Lee, S.-J, Verdine, G.L. | | Deposit date: | 2019-03-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Mechanism of DNA Lesion Homing and Recognition by the Uvr Nucleotide Excision Repair System.
Res, 2019, 2019
|
|
6O8G
 
 | | Crystal structure of UvrB bound to fully duplex DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*CP*GP*AP*TP*GP*GP*AP*GP*A)-3'), ... | | Authors: | Lee, S.-J, Sung, R.-J, Verdine, G.L. | | Deposit date: | 2019-03-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Mechanism of DNA Lesion Homing and Recognition by the Uvr Nucleotide Excision Repair System.
Res, 2019, 2019
|
|
2R6F
 
 | | Crystal Structure of Bacillus stearothermophilus UvrA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Excinuclease ABC subunit A, ZINC ION | | Authors: | Inuzuka, Y, Pakotiprapha, D, Bowman, B.R, Jeruzalmi, D, Verdine, G.L. | | Deposit date: | 2007-09-05 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Bacillus stearothermophilus UvrA Provides Insight into ATP-Modulated Dimerization, UvrB Interaction, and DNA Binding.
Mol.Cell, 29, 2008
|
|
1ADN
 
 | |
1L2D
 
 | | MutM (Fpg)-DNA Estranged Guanine Mismatch Recognition Complex | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*GP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L1Z
 
 | | MutM (Fpg) Covalent-DNA Intermediate | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
1L2B
 
 | | MutM (Fpg) DNA End-Product Structure | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*C*(AD2))-3', 5'-D(P*GP*TP*CP*TP*AP*CP*C)-3', ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
2CO0
 
 | | WDR5 and unmodified Histone H3 complex at 2.25 Angstrom | | Descriptor: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | Authors: | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
2CNX
 
 | | WDR5 and Histone H3 Lysine 4 dimethyl complex at 2.1 angstrom | | Descriptor: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | Authors: | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
1ZXN
 
 | | Human DNA topoisomerase IIa ATPase/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA topoisomerase II, alpha isozyme, ... | | Authors: | Wei, H, Ruthenburg, A.J, Bechis, S.K, Verdine, G.L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nucleotide-dependent Domain Movement in the ATPase Domain of a Human Type IIA DNA Topoisomerase.
J.Biol.Chem., 280, 2005
|
|
