4E1G
 
 | |
7KP4
 
 | |
6OV3
 
 | |
6OV2
 
 | |
8U5B
 
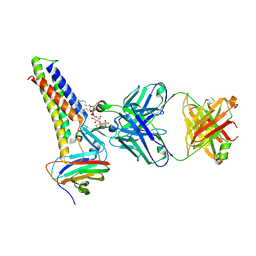 | |
8U4V
 
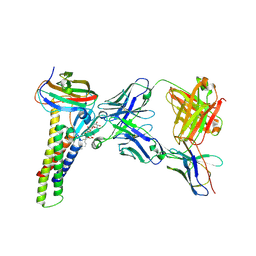 | |
3MDL
 
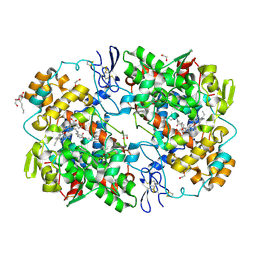
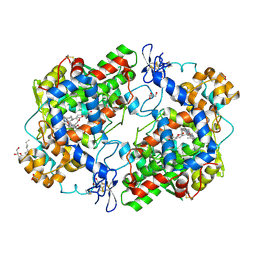 | | X-ray crystal structure of 1-arachidonoyl glycerol bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | (2S)-2,3-dihydroxypropyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Malkowski, M.G. | | Deposit date: | 2010-03-30 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis of endocannabinoid oxygenation by cyclooxygenase-2.
J.Biol.Chem., 286, 2011
|
|
9BEI
 
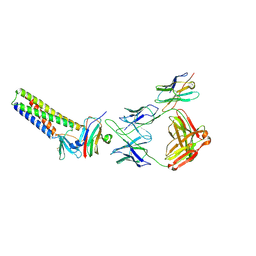 | |
3OLT
 
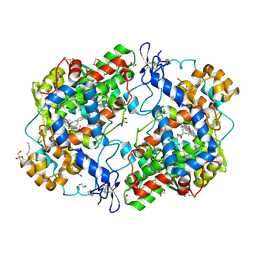 | |
3OLU
 
 | | X-ray crystal structure of 1-arachidonoyl glycerol bound to the cyclooxygenase channel of R513H murine COX-2 | | Descriptor: | (2S)-2,3-dihydroxypropyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Malkowski, M.G. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structural basis of endocannabinoid oxygenation by cyclooxygenase-2.
J.Biol.Chem., 286, 2011
|
|
3HS5
 
 | | X-ray crystal structure of arachidonic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
3HS6
 
 | | X-ray crystal structure of eicosapentaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
3HS7
 
 | | X-ray crystal structure of docosahexaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
3KRK
 
 | | X-ray crystal structure of arachidonic acid bound in the cyclooxygenase channel of L531F murine COX-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-11-18 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
3QH0
 
 | |
3QMO
 
 | |
3TZI
 
 | |
7TDM
 
 | |
7TDN
 
 | |
8U5F
 
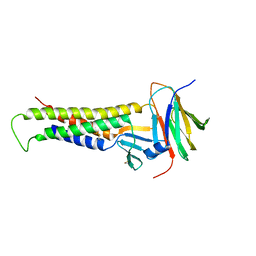
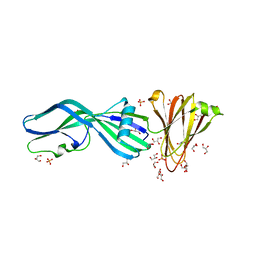 | | Crystal Structure of Trypsinized Clostridium perfringens Enterotoxin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Kapoor, S, Ogbu, C.P, Vecchio, A.J. | | Deposit date: | 2023-09-12 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin.
Toxins, 15, 2023
|
|
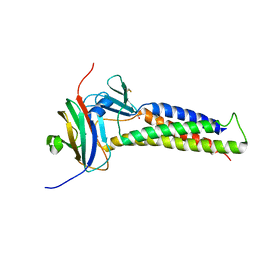
8U5E
 
 | |
8U5D
 
 | |
