8IPA
 
 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently with cycloheximide | | Descriptor: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
2MGZ
 
 | | Solution structure of RBFOX family ASD-1 RRM and SUP-12 RRM in ternary complex with RNA | | Descriptor: | Protein ASD-1, isoform a, Protein SUP-12, ... | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
8IP9
 
 | | Wheat 40S ribosome in complex with a tRNAi | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S23, 40S ribosomal protein eS1, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
5BW7
 
 | | Crystal structure of nonfucosylated Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Isoda, Y, Yagi, H, Satoh, T, Shibata-Koyama, M, Masuda, K, Satoh, M, Kato, K, Iida, S. | | Deposit date: | 2015-06-06 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Importance of the Side Chain at Position 296 of Antibody Fc in Interactions with Fc gamma RIIIa and Other Fc gamma Receptors
Plos One, 10, 2015
|
|
7DJI
 
 | |
4TV7
 
 | |
2RNE
 
 | | Solution structure of the second RNA recognition motif (RRM) of TIA-1 | | Descriptor: | Tia1 protein | | Authors: | Takahashi, M, Kuwasako, K, Abe, C, Tsuda, K, Inoue, M, Terada, T, Shirouzu, M, Kobayashi, N, Kigawa, T, Taguchi, S, Guntert, P, Hayashizaki, Y, Tanaka, A, Muto, Y, Yokoyama, S. | | Deposit date: | 2007-12-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode
Biochemistry, 47, 2008
|
|
4D9T
 
 | | Rsk2 C-terminal Kinase Domain with inhibitor (E)-methyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, methyl (2S)-3-{4-amino-7-[(1E)-3-hydroxyprop-1-en-1-yl]-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl}-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
2RU3
 
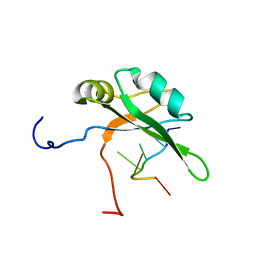 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
1M7W
 
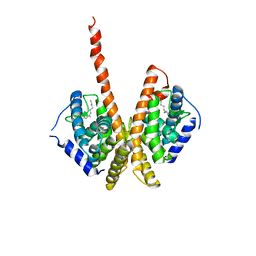 | | HNF4a ligand binding domain with bound fatty acid | | Descriptor: | Hepatocyte nuclear factor 4-alpha, LAURIC ACID | | Authors: | Dhe-Paganon, S, Duda, K, Iwamoto, M, Chi, Y.I, Shoelson, S.E. | | Deposit date: | 2002-07-22 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand
J.Biol.Chem., 277, 2002
|
|
1O5P
 
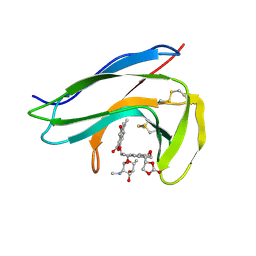 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
4D9U
 
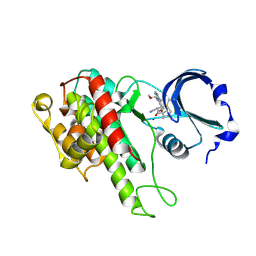 | | Rsk2 C-terminal Kinase Domain, (E)-tert-butyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, tert-butyl (2S)-3-[4-amino-7-(3-hydroxypropyl)-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl]-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
2LKO
 
 | | Structural Basis of Phosphoinositide Binding to Kindlin-2 Pleckstrin Homology Domain in Regulating Integrin Activation | | Descriptor: | Fermitin family homolog 2, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Liu, J, Fukuda, K, Xu, Z. | | Deposit date: | 2011-10-17 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation.
J.Biol.Chem., 286, 2011
|
|
2K2R
 
 | | The NMR structure of alpha-parvin CH2/paxillin LD1 complex | | Descriptor: | Alpha-parvin, Paxillin | | Authors: | Wang, X, Fukuda, K, Byeon, I, Velyvis, A, Wu, C, Gronenborn, A, Qin, J. | | Deposit date: | 2008-04-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of {alpha}-Parvin CH2-Paxillin LD1 Complex Reveals a Novel Modular Recognition for Focal Adhesion Assembly.
J.Biol.Chem., 283, 2008
|
|
1JWI
 
 | | Crystal Structure of Bitiscetin, a von Willeband Factor-dependent Platelet Aggregation Inducer. | | Descriptor: | bitiscetin, platelet aggregation inducer | | Authors: | Hirotsu, S, Mizuno, H, Fukuda, K, Qi, M.C, Matsui, T, Hamako, J, Morita, T, Titani, K. | | Deposit date: | 2001-09-04 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of bitiscetin, a von Willebrand factor-dependent platelet aggregation inducer.
Biochemistry, 40, 2001
|
|
5X7E
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) in complex with 1,25-dihydroxyvitamin D2 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(E,2R,5S)-5,6-dimethyl-6-oxidanyl-hept-3-en-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydr o-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D3 dihydroxylase | | Authors: | Hayashi, K, Yasuda, K, Shiro, Y, Sugimoto, H, Sakaki, T. | | Deposit date: | 2017-02-25 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Production of an active form of vitamin D2 by genetically engineered CYP105A1
Biochem. Biophys. Res. Commun., 486, 2017
|
|
2MWN
 
 | | Talin-F3 / RIAM N-terminal Peptide complex | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, Talin-1 | | Authors: | Yang, J, Zhu, L, Zhang, H, Hirbawi, J, Fukuda, K, Dwivedi, P, Liu, J, Byzova, T, Plow, E.F, Wu, J, Qin, J. | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational activation of talin by RIAM triggers integrin-mediated cell adhesion.
Nat Commun, 5, 2014
|
|
2OTN
 
 | | Crystal structure of the catalytically active form of diaminopimelate epimerase from Bacillus anthracis | | Descriptor: | Diaminopimelate epimerase | | Authors: | Matho, M.H, Fukuda, K, Santelli, E, Jaroszewski, L, Liddington, R.C, Roper, D. | | Deposit date: | 2007-02-08 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and inhibition of a catalytically active form of diaminopimelate epimerase (DapF)from Bacillus anthracis
To be Published
|
|
1MT5
 
 | | CRYSTAL STRUCTURE OF FATTY ACID AMIDE HYDROLASE | | Descriptor: | Fatty-acid amide hydrolase, METHYL ARACHIDONYL FLUOROPHOSPHONATE | | Authors: | Bracey, M.H, Hanson, M.A, Masuda, K.R, Stevens, R.C, Cravatt, B.F. | | Deposit date: | 2002-09-20 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Adaptations in a Membrane Enzyme That Terminates Endocannabinoid Signaling
science, 298, 2002
|
|
1UL1
 
 | | Crystal structure of the human FEN1-PCNA complex | | Descriptor: | Flap endonuclease-1, MAGNESIUM ION, Proliferating cell nuclear antigen | | Authors: | Sakurai, S, Kitano, K, Yamaguchi, H, Hamada, K, Okada, K, Fukuda, K, Uchida, M, Ohtsuka, E, Morioka, H, Hakoshima, T. | | Deposit date: | 2003-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for recruitment of human flap endonuclease 1 to PCNA
EMBO J., 24, 2005
|
|
2YSO
 
 | | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog | | Descriptor: | ZINC ION, Zinc finger protein 95 homolog | | Authors: | Takahashi, M, Kuwasako, K, Tsuda, K, Tanabe, W, Harada, T, Watanabe, S, Tochio, N, Muto, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog
To be Published
|
|
2RPP
 
 | | Solution structure of Tandem zinc finger domain 12 in Muscleblind-like protein 2 | | Descriptor: | Muscleblind-like protein 2, ZINC ION | | Authors: | Abe, C, Dang, W, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
2RT9
 
 | | Solution structure of a regulatory domain of meiosis inhibitor | | Descriptor: | F-box only protein 43, ZINC ION | | Authors: | Shoji, S, Muto, Y, Ikeda, M, He, F, Tsuda, K, Ohsawa, N, Akasaka, R, Terada, T, Wakiyama, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The zinc-binding region (ZBR) fragment of Emi2 can inhibit APC/C by targeting its association with the coactivator Cdc20 and UBE2C-mediated ubiquitylation
FEBS Open Bio, 4, 2014
|
|
2RS2
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | Deposit date: | 2011-06-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
