8C12
 
 | | Identification of an intermediate activation state of PAK5 reveals a novel mechanism of kinase inhibition. | | Descriptor: | PAK5-Af17, Serine/threonine-protein kinase PAK 5 | | Authors: | Martin, H.L, Turner, A.L, Trinh, C.H, Bayliss, R.W, Tomlinson, D.C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Affimer-mediated locking of p21-activated kinase 5 in an intermediate activation state results in kinase inhibition.
Cell Rep, 42, 2023
|
|
6GRR
 
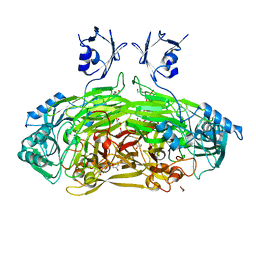 | | Crystal structure of Escherichia coli amine oxidase mutant I342F/E573Q | | Descriptor: | Amine oxidase, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Gaule, T.G, Smith, M.A, Tych, K.M, Pirrat, P, Trinh, C.H, Pearson, A.R, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2018-06-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxygen Activation Switch in the Copper Amine Oxidase of Escherichia coli.
Biochemistry, 57, 2018
|
|
3S4G
 
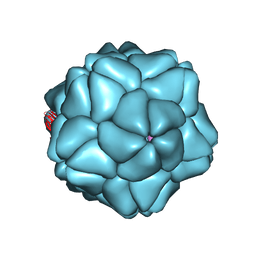 | | Low Resolution Structure of STNV complexed with RNA | | Descriptor: | Capsid protein, RNA (5'-R(P*AP*AP*A)-3'), RNA (5'-R(P*UP*UP*UP*U)-3') | | Authors: | Lane, S.W, Dennis, C.A, Lane, C.L, Trinh, C.H, Rizkallah, P.J, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2011-05-19 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Construction and crystal structure of recombinant STNV capsids.
J.Mol.Biol., 413, 2011
|
|
6HNL
 
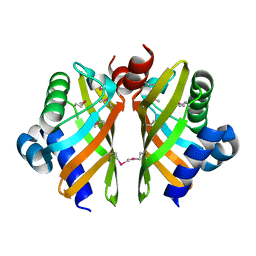 | | Selenomethionine derivative of IdmH 96-104 loop truncation variant | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
6HNM
 
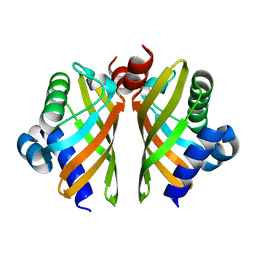 | | Crystal structure of IdmH 96-104 loop truncation variant | | Descriptor: | putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
5ZZ1
 
 | | Probing the active center of catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, CALCIUM ION, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Yuzugullu Karakus, Y, Trinh, C.H, Pearson, A.R, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of the site of oxidase substrate binding in Scytalidium thermophilum catalase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4AHP
 
 | | Crystal Structure of Wild Type N-acetylneuraminic acid lyase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
4AHQ
 
 | | Crystal Structure of N-acetylneuraminic acid lyase mutant K165C from Staphylococcus aureus | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
4AHO
 
 | | Crystal Structure of N-acetylneuraminic acid lyase from Staphylococcus aureus with the chemical modification thia-lysine at position 165 | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
6HNN
 
 | | Crystal structure of wild-type IdmH, a putative polyketide cyclase from Streptomyces antibioticus | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
2Q81
 
 | | Crystal Structure of the Miz-1 BTB/POZ domain | | Descriptor: | Miz-1 protein, TETRAETHYLENE GLYCOL | | Authors: | Stead, M.A, Trinh, C.H, Garnett, J.A, Carr, S.B, Edwards, T.A, Wright, S.C. | | Deposit date: | 2007-06-08 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Beta-Sheet Interaction Interface Directs the Tetramerisation of the Miz-1 POZ Domain
J.Mol.Biol., 373, 2007
|
|
6S0D
 
 | | H30 MnSOD-3 Mutant II | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] 2, ... | | Authors: | Bonetta, R, Trinh, C.H, Hunter, G.J, Hunter, T. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | H30 MnSOD-3 Mutant II
To Be Published
|
|
5EQL
 
 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SUMO-Affirmer-S2D5, Small ubiquitin-related modifier 2 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
5ELU
 
 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SULFATE ION, SUMO-Affirmer-S2B3, Small ubiquitin-related modifier 2 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-05 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
5ELJ
 
 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SUMO-Affirmer-S2D5, Small ubiquitin-related modifier 1 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
6SWT
 
 | |
5IU1
 
 | | N-terminal PAS domain homodimer of PpANR MAP3K from Physcomitrella patens. | | Descriptor: | CTR1-like protein, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stevenson, R.R, Trinh, C.H, Edwards, T.A, Cuming, A.C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Genetic Analysis of Physcomitrella patens Identifies ABSCISIC ACID NON-RESPONSIVE, a Regulator of ABA Responses Unique to Basal Land Plants and Required for Desiccation Tolerance.
Plant Cell, 28, 2016
|
|
5LKY
 
 | | X-ray crystal structure of N-acetylneuraminic acid lyase in complex with pyruvate, with the phenylalanine at position 190 replaced with the non-canonical amino acid dihydroxypropylcysteine. | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetylneuraminate lyase | | Authors: | Windle, C.L, Trinh, C.H, Pearson, A.R, Nelson, A.S, Berry, A. | | Deposit date: | 2016-07-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending enzyme molecular recognition with an expanded amino acid alphabet.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6YWZ
 
 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 2-[(~{E})-(4-oxidanylidenebutanoylhydrazinylidene)methyl]benzoic acid, ARGININE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Lian, L.J, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
6ST2
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Affimer AF6, Bcl-2-like protein 1, SULFATE ION | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
5A37
 
 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | HUMAN ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-05-27 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
5A4B
 
 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | HUMAN ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-06-05 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
6YX0
 
 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 4-[[2-(4-oxidanylidenebutanoyl)hydrazinyl]methyl]benzoic acid, PWQ-THR-ARG-LEU, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
5A36
 
 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-05-27 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
5A38
 
 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-05-27 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
