3P8R
 
 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE FROM Vibrio cholerae | | Descriptor: | Geranyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3RBG
 
 | | Crystal structure analysis of Class-I MHC restricted T-cell associated molecule | | Descriptor: | Cytotoxic and regulatory T-cell molecule, PHOSPHATE ION | | Authors: | Rubinstein, R, Ramagopal, U.A, Toro, R, Nathenson, S.G, Fiser, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-03-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional classification of immune regulatory proteins.
Structure, 21, 2013
|
|
3JU2
 
 | | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein SMc04130 | | Authors: | Patskovsky, Y, Foti, R, Ramagopal, U, Malashkevich, V, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti
To be Published
|
|
3G79
 
 | | Crystal structure of NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase from Methanosarcina mazei Go1 | | Descriptor: | NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase from Methanosarcina mazei Go1
To be Published
|
|
3HPA
 
 | | Crystal structure of an amidohydrolase gi:44264246 from an evironmental sample of sargasso sea | | Descriptor: | AMIDOHYDROLASE, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The hunt for 8-oxoguanine deaminase.
J.Am.Chem.Soc., 132, 2010
|
|
4FP4
 
 | | Crystal structure of isoprenoid synthase a3mx09 (target efi-501993) from pyrobaculum calidifontis | | Descriptor: | GERAN-8-YL GERAN, Polyprenyl synthetase, UNKNOWN ATOM OR ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GI5
 
 | | Crystal Structure Of a Putative quinone reductase from Klebsiella pneumoniae (Target PSI-013613) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a quinone reductase from Klebsiella pneumoniae with bound FAD
to be published
|
|
4GEK
 
 | | Crystal Structure of wild-type CmoA from E.coli | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, SULFATE ION, tRNA (cmo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bonanno, J.B, Bhosle, R, Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Nature, 498, 2013
|
|
4DI8
 
 | | CRYSTAL STRUCTURE OF THE D248A mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 8.5 | | Descriptor: | (1E,3Z)-4-hydroxybuta-1,3-diene-1,2,4-tricarboxylic acid, 2-oxo-2H-pyran-4,6-dicarboxylic acid, 2-pyrone-4,6-dicarbaxylate hydrolase, ... | | Authors: | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4E8G
 
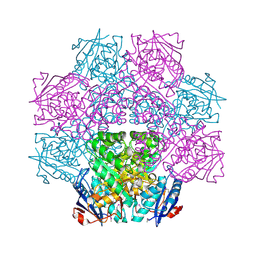 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound mg | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
4GOS
 
 | | Crystal structure of human B7-H4 IgV-like domain | | Descriptor: | V-set domain-containing T-cell activation inhibitor 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Vigdorovich, V, Ramagopal, U, Bhosle, R, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and cancer immunotherapy of the B7 family member B7x.
Cell Rep, 9, 2014
|
|
4GIC
 
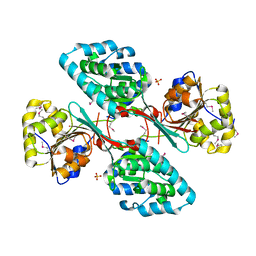 | | Crystal Structure Of a Putative Histidinol dehydrogenase (Target PSI-014034) from Methylococcus capsulatus | | Descriptor: | Histidinol dehydrogenase, SULFATE ION | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a putative Histidinol dehydrogenase from Methylococcus capsulatus
to be published
|
|
4I0K
 
 | | Crystal structure of murine B7-H3 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD276 antigen, SULFATE ION, ... | | Authors: | Vigdorovich, V, Ramagopal, U, Bonanno, J.B, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structure and T cell inhibition properties of B7 family member, B7-H3.
Structure, 21, 2013
|
|
4HWN
 
 | | Crystal structure of the second Ig-C2 domain of human Fc-receptor like A (FCRLA), Isoform 9 [NYSGRC-005836] | | Descriptor: | Fc receptor-like A | | Authors: | Kumar, P.R, Ahmed, M, Bhosle, R, Calarese, D, Celikigil, A, Chan, M.K, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Rubinstein, R, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-08 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of the second Ig-C2 domain of the human Fc-receptor like A, Isoform 9 [NYSGRC-005836]
to be published
|
|
4HWU
 
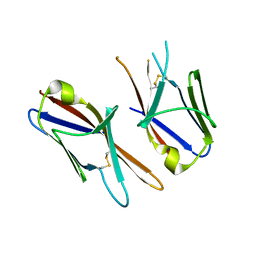 | | Crystal structure of the Ig-C2 type 1 domain from mouse Fibroblast growth factor receptor 2 (FGFR2) [NYSGRC-005912] | | Descriptor: | Fibroblast growth factor receptor 2 | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Rubinstein, R, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of the Ig-C2 type 1 domain from mouse FGFR2 [NYSGRC-005912]
to be published
|
|
4J1O
 
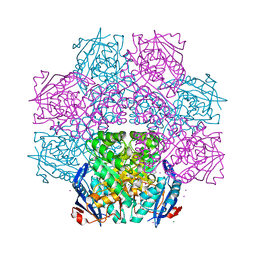 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound l-proline betaine (substrate) | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, GLYCEROL, IODIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
4KNP
 
 | | Crystal Structure Of a Putative enoyl-coA hydratase (PSI-NYSGRC-019597) from Mycobacterium avium paratuberculosis K-10 | | Descriptor: | enoyl-CoA hydratase | | Authors: | Kumar, P.R, Ahmed, M, Attonito, J, Bhosle, R, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-10 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of a Putative enoyl-coA hydratase from Mycobacterium avium paratuberculosis K-10
to be published
|
|
4IZG
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound cis-4oh-d-proline betaine (product) | | Descriptor: | (2S,4S)-2-carboxy-4-hydroxy-1,1-dimethylpyrrolidinium, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
4F62
 
 | | Crystal structure of a putative farnesyl-diphosphate synthase from Marinomonas sp. MED121 (Target EFI-501980) | | Descriptor: | CHLORIDE ION, GLYCEROL, Geranyltranstransferase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LN5
 
 | | Crystal structure of a trap periplasmic solute binding protein from burkholderia ambifaria (Bamb_6123), TARGET EFI-510059, with bound glycerol and chloride ion | | Descriptor: | CHLORIDE ION, GLYCEROL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4KGQ
 
 | | Crystal structure of a human light loop mutant in complex with dcr3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Sampathkumar, P, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-29 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4KKN
 
 | | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704 | | Descriptor: | Cytotoxic T-lymphocyte associated protein 4, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Bonanno, J, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Scott Glenn, A, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704
to be published
|
|
4MHF
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Glucuronate, space group P21 | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit, alpha-D-glucopyranuronic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4KTO
 
 | | Crystal Structure Of a Putative Isovaleryl-CoA dehydrogenase (PSI-NYSGRC-012251) from Sinorhizobium meliloti 1021 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, isovaleryl-CoA dehydrogenase | | Authors: | Kumar, P.R, Ahmed, M, Attonito, J, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Himmel, D, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Crystal structure of a Putative Isovaleryl-CoA dehydrogenase from Sinorhizobium meliloti 1021
to be published
|
|
4KGG
 
 | | Crystal structure of light mutant2 and dcr3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Bonanno, J.B, Zhan, C, Kumar, P.R, Toro, R, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
