6ML4
 
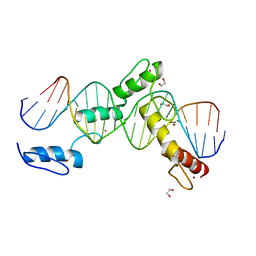 | | BTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 3) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML2
 
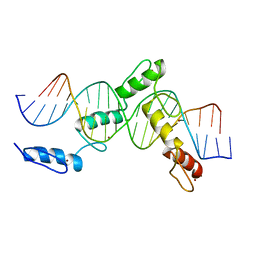 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 1) | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML7
 
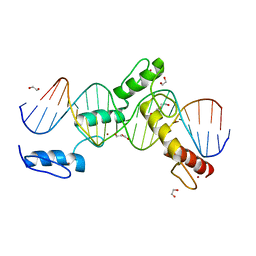 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpG 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*(5CM)P*GP*AP*AP*TP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML3
 
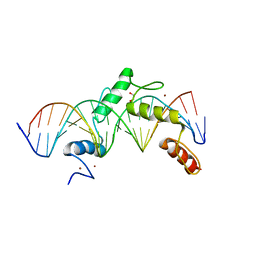 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 2) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
7N5U
 
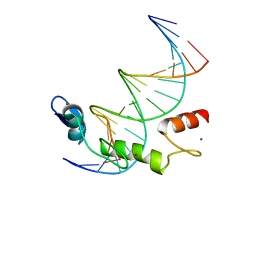 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 21) | | Descriptor: | DNA Strain II, DNA Strand I, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5W
 
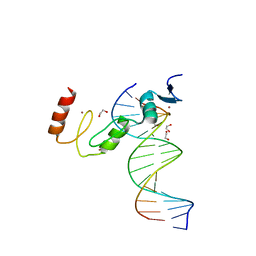 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 23) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand I, DNA Strand II, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5V
 
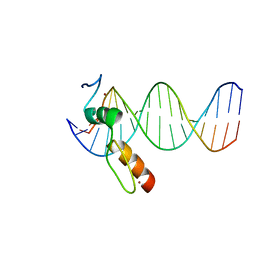 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
6NV1
 
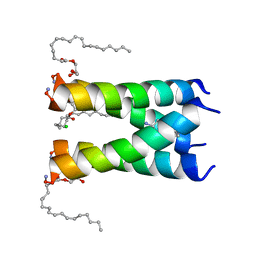 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, ... | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-02-04 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
6O04
 
 | | M.tb MenD IntII bound with Inhibitor | | Descriptor: | (1~{R},2~{S},5~{S},6~{S})-2-[(1~{S})-1-[3-[(4-azanylidene-2-methyl-1~{H}-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl (phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3-ium-2-yl]-1,4-bis(oxidanyl)-4-oxidanylidene-butyl]-6-oxidanyl-5-(3-oxid anyl-3-oxidanylidene-prop-1-en-2-yl)oxy-cyclohex-3-ene-1-carboxylic acid, 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M, Jirgis, E.M.N, Nigon, L.V, Chuang, H, Baker, E.N. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
6O0G
 
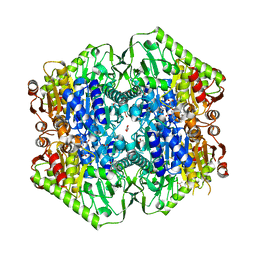 | | M.tb MenD bound to Intermediate I and Inhibitor | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-OXOGLUTARIC ACID, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M.M, Jirgis, E.M.N, Chuang, H, Nigon, L.V, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
6O0N
 
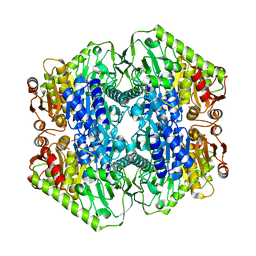 | | M.tb MenD with Inhibitor | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Johnston, J.M, Ho, N.A.T, Bashiri, G, Bulloch, E.M, Nigon, L.V, Jirgis, E.M.N, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
6ZPG
 
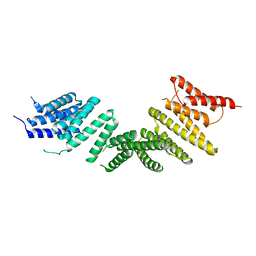 | | Kinesin binding protein (KBP) | | Descriptor: | KIF-binding protein | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
6ZPI
 
 | | Microtubule complexed with Kif15 motor domain. Symmetrised asymmetric unit | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF15, ... | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
4TWS
 
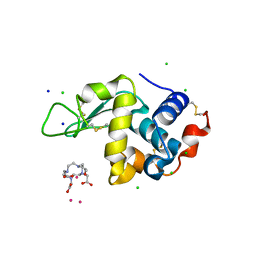 | | Gadolinium Derivative of Tetragonal Hen Egg-White Lysozyme at 1.45 A Resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Holton, J.M, Classen, S, Frankel, K.A, Tainer, J.A. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The R-factor gap in macromolecular crystallography: an untapped potential for insights on accurate structures.
Febs J., 281, 2014
|
|
6O0J
 
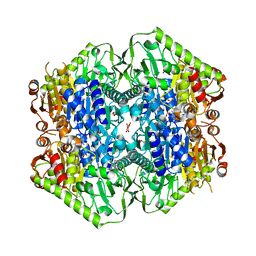 | | M.tb MenD with ThDP and Inhibitor bound | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ACETATE ION, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M.M, Jirgis, E.M.N, Nigon, L.V, Chuang, H, Ho, N.A.T, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
6OD5
 
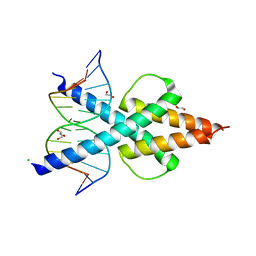 | | Human TCF4 C-terminal bHLH domain in Complex with 12-bp Oligonucleotide Containing E-box Sequence with 5-carboxylcytosines | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*(1CC)P*GP*CP*AP*CP*GP*TP*GP*(1CC)P*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
4UXY
 
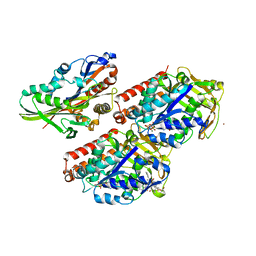 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | ALPHA TUBULIN, BETA TUBULIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
6MNI
 
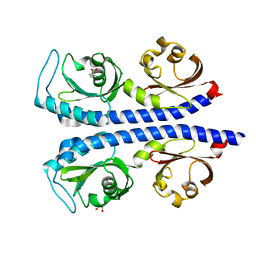 | | Structure of the tandem CACHE domain of PscC | | Descriptor: | CHLORIDE ION, GLYCEROL, Methyl-accepting chemotaxis protein, ... | | Authors: | Johnston, J.M, Gerth, M.L, Ehrhardt, M.K.G. | | Deposit date: | 2018-10-01 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structure of a double CACHE chemoreceptor ligand-binding domain from Pseudomonas syringae provides insights into the basis of proline recognition.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
4RNW
 
 | | Truncated version of the G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RNX
 
 | | K154 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1 | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
7N5T
 
 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 5) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
7N5S
 
 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 6) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
4RNU
 
 | | G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, PHOSPHATE ION | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RNV
 
 | | G303 Circular Permutation of Old Yellow Enzyme with the Inhibitor p-Hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, P-HYDROXYBENZALDEHYDE | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
6OX4
 
 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
