8SVF
 
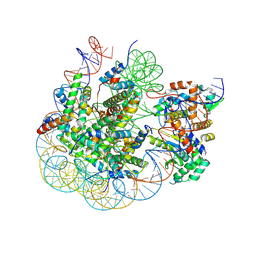 | | BAP1/ASXL1 bound to the H2AK119Ub Nucleosome | | Descriptor: | DNA/RNA (187-MER), DNA/RNA (327-MER), Histone H2A type 1, ... | | Authors: | Thomas, J.F, Valencia-Sanchez, M.I, Armache, K.-J. | | Deposit date: | 2023-05-16 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of histone H2A lysine 119 deubiquitination by Polycomb repressive deubiquitinase BAP1/ASXL1.
Sci Adv, 9, 2023
|
|
3ZNG
 
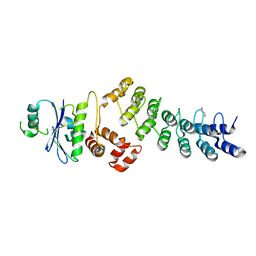 | | Ankyrin repeat and SOCS-box protein 9 (ASB9) in complex with ElonginB and ElonginC | | Descriptor: | 1,2-ETHANEDIOL, ANKYRIN REPEAT AND SOCS BOX PROTEIN 9, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Thomas, J, Van Molle, I, Ciulli, A. | | Deposit date: | 2013-02-14 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Multimeric Complexes Among Ankyrin-Repeat and Socs-Box Protein 9 (Asb9), Elonginbc, and Cullin 5: Insights Into the Structure and Assembly of Ecs-Type Cullin-Ring E3 Ubiquitin Ligases.
Biochemistry, 52, 2013
|
|
1AAB
 
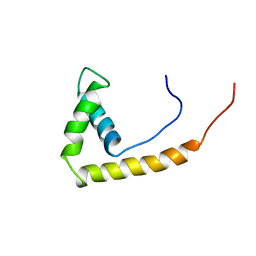 | | NMR STRUCTURE OF RAT HMG1 HMGA FRAGMENT | | Descriptor: | HIGH MOBILITY GROUP PROTEIN | | Authors: | Hardman, C.H, Broadhurst, R.W, Raine, A.R.C, Grasser, K.D, Thomas, J.O, Laue, E.D. | | Deposit date: | 1995-10-28 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 34, 1995
|
|
1YQA
 
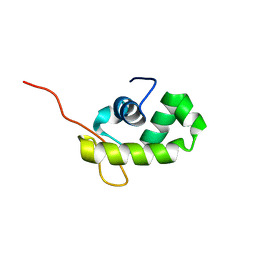 | |
1FLJ
 
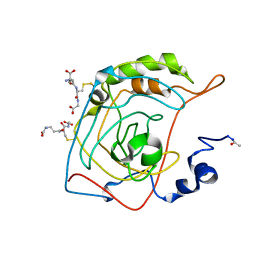 | | CRYSTAL STRUCTURE OF S-GLUTATHIOLATED CARBONIC ANHYDRASE III | | Descriptor: | CARBONIC ANHYDRASE III, GLUTATHIONE, ZINC ION | | Authors: | Mallis, R.J, Poland, B.W, Chatterjee, T.K, Fisher, R.A, Darmawan, S, Honzatko, R.B, Thomas, J.A. | | Deposit date: | 2000-08-14 | | Release date: | 2000-09-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of S-glutathiolated carbonic anhydrase III.
FEBS Lett., 482, 2000
|
|
4N5B
 
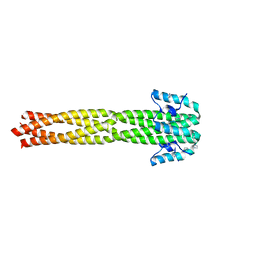 | | Crystal structure of the Nipah virus phosphoprotein tetramerization domain | | Descriptor: | IMIDAZOLE, Phosphoprotein | | Authors: | Bruhn, J.F, Barnett, K, Bibby, J, Thomas, J, Keegan, R, Rigden, D, Bornholdt, Z.A, Saphire, E.O. | | Deposit date: | 2013-10-09 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the nipah virus phosphoprotein tetramerization domain.
J.Virol., 88, 2014
|
|
6HYT
 
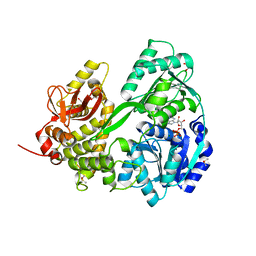 | | Crystal structure of DHX8 helicase domain bound to ADP at 2.3 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
6HYS
 
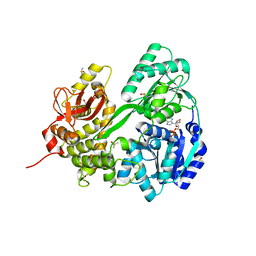 | | Crystal structure of DHX8 helicase domain bound to ADP at 2.6 angstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
1UST
 
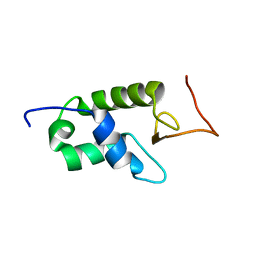 | | YEAST HISTONE H1 GLOBULAR DOMAIN I, HHO1P GI, SOLUTION NMR STRUCTURES | | Descriptor: | HISTONE H1 | | Authors: | Ali, T, Coles, P, Stevens, T.J, Stott, K, Thomas, J.O. | | Deposit date: | 2003-11-30 | | Release date: | 2004-04-01 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Two Homologous Domains of Similar Structure But Different Stability in the Yeast Linker Histone, Hho1P
J.Mol.Biol., 338, 2004
|
|
1USS
 
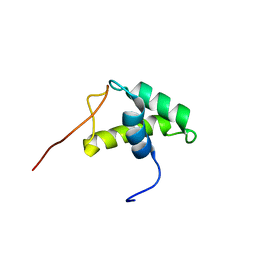 | | YEAST HISTONE H1 GLOBULAR DOMAIN II, HHO1P GII, SOLUTION NMR STRUCTURES | | Descriptor: | HISTONE H1 | | Authors: | Ali, T, Coles, P, Stevens, T.J, Stott, K, Thomas, J.O. | | Deposit date: | 2003-11-30 | | Release date: | 2004-04-01 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Two Homologous Domains of Similar Structure But Different Stability in the Yeast Linker Histone, Hho1P
J.Mol.Biol., 338, 2004
|
|
6HYU
 
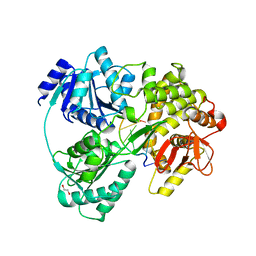 | | Crystal structure of DHX8 helicase bound to single stranded poly-adenine RNA | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DHX8, RNA (5'-R(*A*AP*A)-3'), ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
2UZR
 
 | | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer (AKT1-PH_E17K) | | Descriptor: | RAC-alpha serine/threonine-protein kinase | | Authors: | Carpten, J.D, Faber, A.L, Horn, C, Donoho, G.P, Briggs, S.L, Robbins, C.M, Hostetter, G, Boguslawski, S, Moses, T.Y, Savage, S, Uhlik, M, Lin, A, Du, J, Qian, Y.W, Zeckner, D.J, Tucker-Kellogg, G, Touchman, J, Patel, K, Mousses, S, Bittner, M, Schevitz, R, Lai, M.H, Blanchard, K.L, Thomas, J.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.
Nature, 448, 2007
|
|
2UZS
 
 | | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer (AKT1-PH_E17K) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-alpha serine/threonine-protein kinase | | Authors: | Carpten, J.D, Faber, A.L, Horn, C, Donoho, G.P, Briggs, S.L, Robbins, C.M, Hostetter, G, Boguslawski, S, Moses, T.Y, Savage, S, Uhlik, M, Lin, A, Du, J, Qian, Y.W, Zeckner, D.J, Tucker-Kellogg, G, Touchman, J, Patel, K, Mousses, S, Bittner, M, Schevitz, R, Lai, M.H, Blanchard, K.L, Thomas, J.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.
Nature, 448, 2007
|
|
2GZK
 
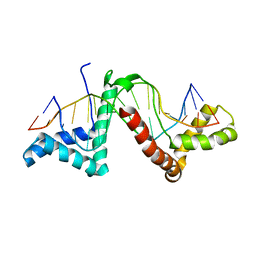 | | Structure of a complex of tandem HMG boxes and DNA | | Descriptor: | 5'-D(*GP*CP*AP*TP*TP*GP*TP*TP*TP*AP*GP*AP*TP*CP*CP*C)-3', 5'-D(*GP*GP*GP*AP*TP*CP*TP*AP*AP*AP*CP*AP*AP*TP*GP*C)-3', Sex-determining region on Y / HMGB1 | | Authors: | Stott, K, Tang, G.S, Lee, K.B, Thomas, J.O. | | Deposit date: | 2006-05-11 | | Release date: | 2006-07-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of a Complex of Tandem HMG Boxes and DNA.
J.Mol.Biol., 360, 2006
|
|
1HME
 
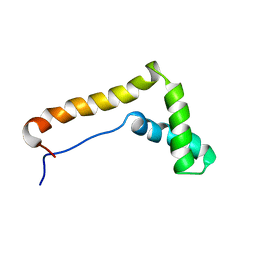 | | STRUCTURE OF THE HMG BOX MOTIF IN THE B-DOMAIN OF HMG1 | | Descriptor: | HIGH MOBILITY GROUP PROTEIN FRAGMENT-B | | Authors: | Weir, H.M, Kraulis, P.J, Hill, C.S, Raine, A.R.C, Laue, E.D, Thomas, J.O. | | Deposit date: | 1994-02-10 | | Release date: | 1994-05-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the HMG box motif in the B-domain of HMG1.
EMBO J., 12, 1993
|
|
2MCO
 
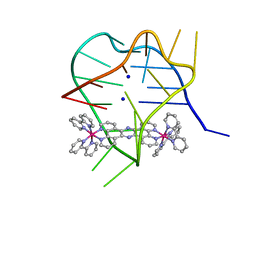 | | Structural studies on dinuclear ruthenium(II) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence | | Descriptor: | SODIUM ION, human telomere quadruplex, tetrakis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)(mu-tetrapyrido[3,2-a:2',3'-c:3'',2''-h:2''',3'''-j]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~13~,N~14~)diruthenium(4+) L enantiomer | | Authors: | Williamson, M.P, Wilson, T, Thomas, J.A, Felix, V, Costa, P.J. | | Deposit date: | 2013-08-22 | | Release date: | 2013-10-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence.
J.Med.Chem., 56, 2013
|
|
2MCC
 
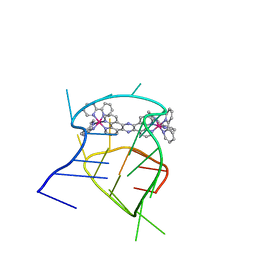 | | Structural studies on dinuclear ruthenium(II) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence | | Descriptor: | human_telomere_quadruplex, tetrakis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)(mu-tetrapyrido[3,2-a:2',3'-c:3'',2''-h:2''',3'''-j]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~13~,N~14~)diruthenium(4+) | | Authors: | Williamson, M.P, Wilson, T, Thomas, J.A, Felix, V, Costa, P.J. | | Deposit date: | 2013-08-18 | | Release date: | 2013-10-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence.
J.Med.Chem., 56, 2013
|
|
1HMF
 
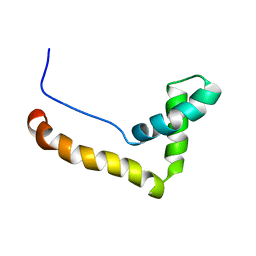 | | STRUCTURE OF THE HMG BOX MOTIF IN THE B-DOMAIN OF HMG1 | | Descriptor: | HIGH MOBILITY GROUP PROTEIN FRAGMENT-B | | Authors: | Weir, H.M, Kraulis, P.J, Hill, C.S, Raine, A.R.C, Laue, E.D, Thomas, J.O. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the HMG box motif in the B-domain of HMG1.
EMBO J., 12, 1993
|
|
2LY4
 
 | | HMGB1-facilitated p53 DNA binding occurs via HMG-box/p53 transactivation domain interaction and is regulated by the acidic tail | | Descriptor: | Cellular tumor antigen p53, High mobility group protein B1 | | Authors: | Rowell, J.P, Simpson, K.L, Stott, K, Watson, M, Thomas, J.O. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-31 | | Last modified: | 2012-12-26 | | Method: | SOLUTION NMR | | Cite: | HMGB1-Facilitated p53 DNA Binding Occurs via HMG-Box/p53 Transactivation Domain Interaction, Regulated by the Acidic Tail.
Structure, 20, 2012
|
|
4AOM
 
 | | MTIP and MyoA complex | | Descriptor: | MYOSIN A TAIL DOMAIN INTERACTING PROTEIN, MYOSIN-A | | Authors: | Salgado, P.S, Douse, C.H, Simpson, P.J, Thomas, J.C, Holder, A.A, Tate, E.W, Cota, E. | | Deposit date: | 2012-03-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Regulation of the Plasmodium Motor Complex: Phosphorylation of Myosin a Tail Interacting Protein (Mtip) Loosens its Grip on Myoa
J.Biol.Chem., 287, 2012
|
|
6SKF
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-08-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
6SKG
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome in TkNat10 deleted strain | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-08-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
6TH6
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-11-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
