8SJK
 
 | | Pembrolizumab Caffeine crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY HEAVY CHAIN, ANTIBODY LIGHT CHAIN, ... | | Authors: | Larpent, P, Codan, L, Bothe, J.R, Stueber, D, Reichert, P, Fischmann, T, Su, Y, Pabit, S, Gupta, S, Iuzzolino, L, Cote, A. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Small-Angle X-ray Scattering as a Powerful Tool for Phase and Crystallinity Assessment of Monoclonal Antibody Crystallites in Support of Batch Crystallization.
Mol Pharm., 21, 2024
|
|
4TXC
 
 | | Crystal Structure of DAPK1 kinase domain in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-{3-[(R)-{[2-(dimethylamino)ethyl]amino}(hydroxy)methyl]phenyl}imidazo[1,2-b]pyridazin-6-yl)-2-methoxyphenol, Death-associated protein kinase 1 | | Authors: | Sorrell, F.J, Krojer, T, Wilbek, T.S, Skovgaard, T, Berthelsen, J, Dixon-Clarke, S, Chalk, R, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Stromgaard, K, Knapp, S. | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Identification and characterization of a small-molecule inhibitor of death-associated protein kinase 1.
Chembiochem, 16, 2015
|
|
6QFS
 
 | | Chargeless variant of the Cellulose-binding domain from Cellulomonas fimi | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ... | | Authors: | Young, D.R, Hoejgaard, C, Messens, J, Winther, J.R. | | Deposit date: | 2019-01-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Charge Interactions in a Highly Charge-depleted Protein
J.Am.Chem.Soc., 2021
|
|
6RIO
 
 | | Imidazole Polyamide-DNA complex NMR structure (5'-CGATGTACATCG-3') | | Descriptor: | 3-[3-[[4-[[4-[[4-[[4-[[(2~{R})-2-azaniumyl-4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]butanoyl]amino]-1-methyl-imidazol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]propanoylamino]propyl-dimethyl-azanium, DNA (5'-(*(DC5)P*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*(DG3))-3') | | Authors: | Padroni, G, Withers, J.M, Taladriz-Sender, A, Reichenbach, L.F, Parkinson, J.A, Burley, G.A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequence-Selective Minor Groove Recognition of a DNA Duplex Containing Synthetic Genetic Components.
J.Am.Chem.Soc., 141, 2019
|
|
6T64
 
 | | A model of the EIAV CA-SP hexamer (C6) from Gag-deltaMA spheres assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
4ZG8
 
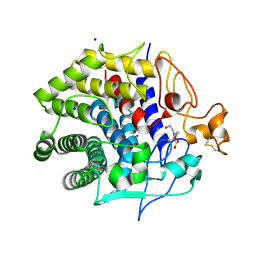 | | Crystal structure of Endoglucanase from Perinereis brevicirris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fewings, R.S, Swiderska, A, Sanchez-Weatherby, J, Sorensen, T.L.-M, Schnorr, K.M, Kneale, G.G, McGeehan, J.E. | | Deposit date: | 2015-04-22 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biophysical and structural characterisation of the endoglucanase from Perinereis brevicirris
To Be Published
|
|
8RQ8
 
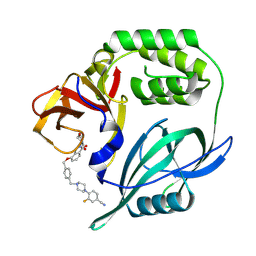 | | Crystal structure of CRBN-midi in complex with mezigdomide | | Descriptor: | Mezigdomide, Protein cereblon, ZINC ION | | Authors: | Zollman, D, Kroupova, A, Pethe, J, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
6T61
 
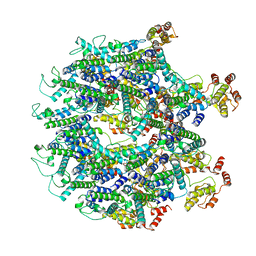 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH8 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6T63
 
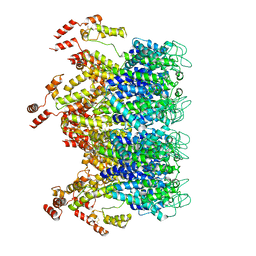 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
1J8V
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4'-nitrophenyl 3I-thiolaminaritrioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Fairweather, J.K, Driguez, H, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2001-05-22 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for broad substrate specificity in higher plant beta-D-glucan glucohydrolases.
Plant Cell, 14, 2002
|
|
1DP5
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT INHIBITOR | | Descriptor: | PROTEINASE A, PROTEINASE INHIBITOR IA3, beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-23 | | Release date: | 2000-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
5N5N
 
 | | Cryo-EM structure of tsA201 cell alpha1B and betaI and betaIVb microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Vemu, A, Atherton, J, Spector, J.O, Moores, C.A, Roll-Mecak, A. | | Deposit date: | 2017-02-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tubulin isoform composition tunes microtubule dynamics.
Mol. Biol. Cell, 28, 2017
|
|
5M54
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-J, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5M50
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 3, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5M5C
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1DPJ
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH IA3 PEPTIDE INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEINASE A, PROTEINASE INHIBITOR IA3 PEPTIDE, ... | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-27 | | Release date: | 2000-05-03 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
5FUF
 
 | | Crystal structure of the Mep2 mutant S453D from Candida albicans | | Descriptor: | AMMONIUM TRANSPORTER, DECYL-BETA-D-MALTOPYRANOSIDE | | Authors: | van den Berg, B, Chembath, A, Jefferies, D, Basle, A, Khalid, S, Rutherford, J. | | Deposit date: | 2016-01-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Structural Basis for Mep2 Ammonium Transceptor Activation by Phosphorylation.
Nat.Commun., 7, 2016
|
|
1KLA
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 1-17 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KLC
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KLD
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 18-33 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
7K7H
 
 | | Density-fitted Model Structure of Antibody Variable Domains of TyTx1 in Complex with PltB pentamer of Typhoid Toxin | | Descriptor: | Fab Heavy Chain Variable Domain, Fab Light Chain Variable Domain, Pertussis like toxin subunit B, ... | | Authors: | Nguyen, T, Feathers, J.R, Fromme, J.C, Song, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The structural basis of Salmonella A 2 B 5 toxin neutralization by antibodies targeting the glycan-receptor binding subunits.
Cell Rep, 36, 2021
|
|
7K7I
 
 | | Density-fitted Model Structure of Antibody Variable Domains of TyTx4 in Complex with PltB pentamer of Typhoid Toxin | | Descriptor: | Fab Heavy Chain Variable Domain, Fab Light Chain Variable Domain, Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Feathers, J.R, Fromme, J.C, Song, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | The structural basis of Salmonella A 2 B 5 toxin neutralization by antibodies targeting the glycan-receptor binding subunits.
Cell Rep, 36, 2021
|
|
6CME
 
 | | Structure of wild-type ISL2-LID in complex with LHX4-LIM1+2 | | Descriptor: | LIM/homeobox protein Lhx4,Insulin gene enhancer protein ISL-2, ZINC ION | | Authors: | Stokes, P.H, Silva, A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins, 87, 2019
|
|
3MMK
 
 | | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification | | Descriptor: | CHLORIDE ION, Fusion of LIM/homeobox protein Lhx4, linker, ... | | Authors: | Gadd, M.S, Langley, D.B, Guss, J.M, Matthews, J.M. | | Deposit date: | 2010-04-20 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification.
J.Biol.Chem., 2011
|
|
4LTJ
 
 | |
