8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
2PJR
 
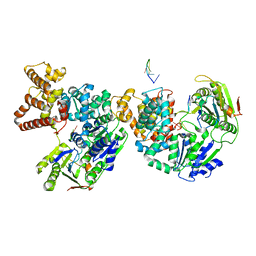 | | HELICASE PRODUCT COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*T)-3'), ... | | Authors: | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
7RPM
 
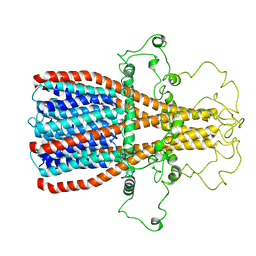 | |
4Z91
 
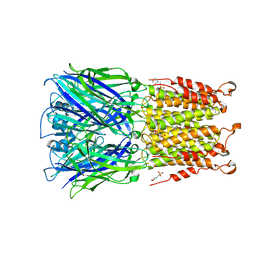 | | ELIC cocrystallized with isofluorane in a desensitized state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 1.7.6 3-bromanylpropan-1-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Chen, Q, Kinde, M.N, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3915 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
1G58
 
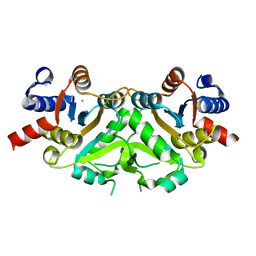 | | CRYSTAL STRUCTURE OF 3,4-DIHYDROXY-2-BUTANONE 4-PHOSPHATE SYNTHASE GOLD DERIVATIVE | | Descriptor: | 3,4-DIHYDROXY-2-BUTANONE 4-PHOSPHATE SYNTHASE, GOLD ION | | Authors: | Liao, D.-I, Calabrese, J.C, Wawrzak, Z, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2000-10-30 | | Release date: | 2001-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase of riboflavin biosynthesis.
Structure, 9, 2001
|
|
1B50
 
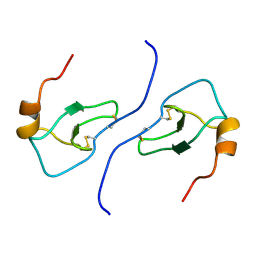 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, 10 STRUCTURES | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
1B53
 
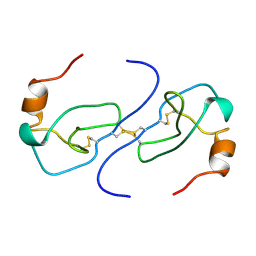 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
6IZT
 
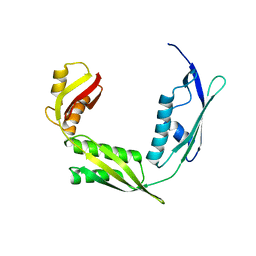 | | Crystal structure of Haemophilus Influenzae BamA POTRA3-5 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6IZS
 
 | | Crystal structure of Haemophilus influenzae BamA POTRA4 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6J09
 
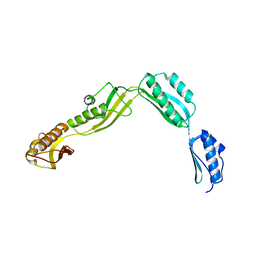 | | Crystal structure of Haemophilus Influenzae BamA POTRA1-4 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | Deposit date: | 2018-12-21 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
7NNL
 
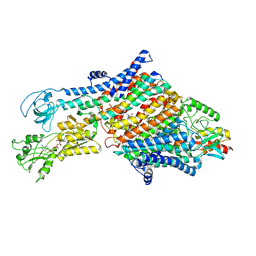 | | Cryo-EM structure of the KdpFABC complex in an E1-ATP conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
7NNP
 
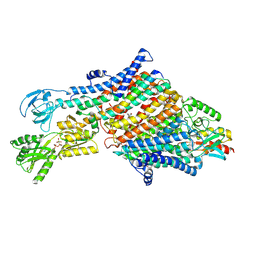 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
8CTH
 
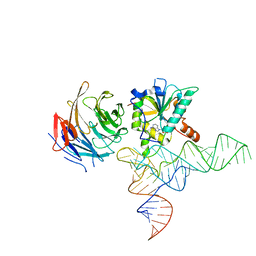 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Phe) complex | | Descriptor: | Phe-tRNA, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(7)-)-methyltransferase, ... | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fishcer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
8CTI
 
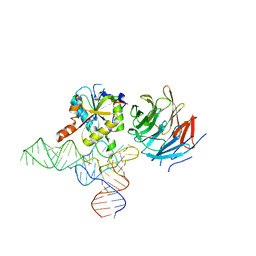 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Val) complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4, tRNA-Val-TAC-2-1 | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fischer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
7TPJ
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7TPG
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
4Z90
 
 | | ELIC bound with the anesthetic isoflurane in the resting state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Chen, Q, Kinde, M, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
8OLR
 
 | | Structure of yeast 20S proteasome in complex with the natural product beta-lactone inhibitor Cystargolide A | | Descriptor: | CHLORIDE ION, Cystargolide A (bound), MAGNESIUM ION, ... | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OLL
 
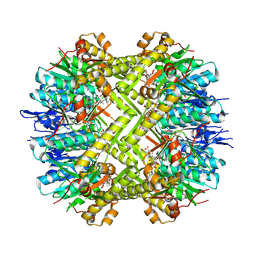 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.7 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6UJ8
 
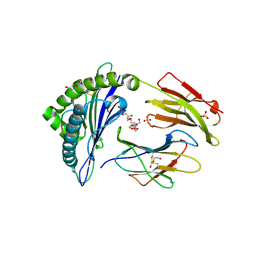 | | Crystal structure of HLA-B*07:02 with wild-type IDH2 peptide | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, HLA class I histocompatibility antigen, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6UJ9
 
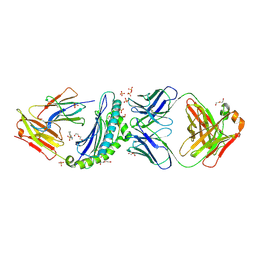 | | Crystal structure of HLA-B*07:02 with R140Q mutant IDH2 peptide in complex with Fab | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Thirawatananond, P, Aytenfisu, T.Y, Wright, K, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6UJ7
 
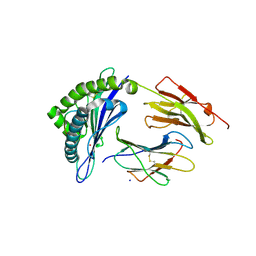 | | Crystal structure of HLA-B*07:02 with R140Q mutant IDH2 peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
7ZRK
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
6XKK
 
 | | Cryo-EM structure of the NLRP1-CARD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | Deposit date: | 2020-06-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
6X6C
 
 | | Cryo-EM structure of NLRP1-DPP9-VbP complex | | Descriptor: | Dipeptidyl peptidase 9, NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Hollingsworth, L.R, Sharif, H, Griswold, A.R, Fontana, P, Mintseris, J, Dagbay, K.B, Paulo, J.A, Gygi, S.P, Bachovchin, D.A, Wu, H. | | Deposit date: | 2020-05-27 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation.
Nature, 592, 2021
|
|
