3LZB
 
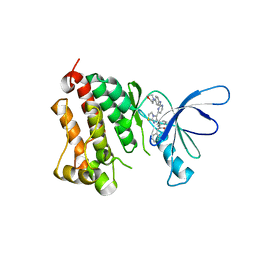 | | EGFR kinase domain complexed with an imidazo[2,1-b]thiazole inhibitor | | Descriptor: | Epidermal growth factor receptor, N-[3-(5-{2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl}imidazo[2,1-b][1,3]thiazol-6-yl)phenyl]-2-phenylacetamide | | Authors: | Swinger, K.K. | | Deposit date: | 2010-03-01 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Imidazo[2,1-b]thiazoles: multitargeted inhibitors of both the insulin-like growth factor receptor and members of the epidermal growth factor family of receptor tyrosine kinases.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2HT0
 
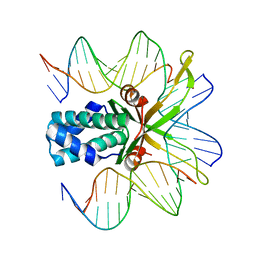 | | IHF bound to doubly nicked DNA | | Descriptor: | 5'-D(*CP*GP*GP*TP*GP*CP*AP*AP*CP*AP*AP*AP*T)-3', 5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3', 5'-D(*GP*GP*CP*CP*AP*AP*AP*AP*AP*AP*GP*CP*AP*TP*T)-3', ... | | Authors: | Swinger, K.K, Rice, P.A. | | Deposit date: | 2006-07-24 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based Analysis of HU-DNA Binding.
J.Mol.Biol., 365, 2007
|
|
6WE3
 
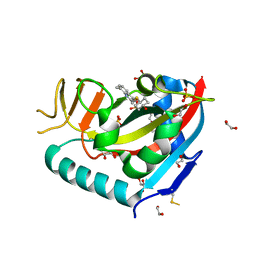 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}-8-methylquinazolin-4(3H)-one, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
6WE4
 
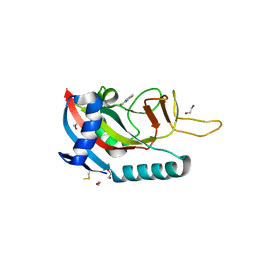 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one, 8-methyl-2-{[(pyridin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, ... | | Authors: | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
6V3W
 
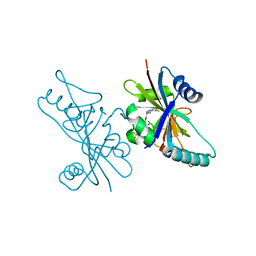 | | Human Poly(ADP-Ribose) Polymerase 12, Catalytic fragment with four point mutations in complex with RBN-2397 | | Descriptor: | 5-{[(2S)-1-(3-oxo-3-{4-[5-(trifluoromethyl)pyrimidin-2-yl]piperazin-1-yl}propoxy)propan-2-yl]amino}-4-(trifluoromethyl)pyridazin-3(2H)-one, CHLORIDE ION, Protein mono-ADP-ribosyltransferase PARP12 | | Authors: | Swinger, K.K, Gozgit, J.M, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity.
Cancer Cell, 39, 2021
|
|
6W65
 
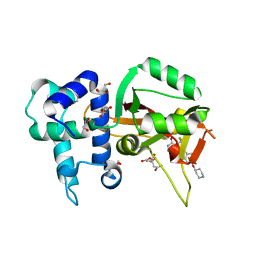 | | Human PARP16 in complex with RBN010860 | | Descriptor: | 1,2-ETHANEDIOL, 5-{5-[(piperidin-4-yl)oxy]-2H-isoindol-2-yl}-4-(trifluoromethyl)pyridazin-3(2H)-one, CITRIC ACID, ... | | Authors: | Swinger, K.K, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | In Vitro and Cellular Probes to Study PARP Enzyme Target Engagement.
Cell Chem Biol, 27, 2020
|
|
6WE2
 
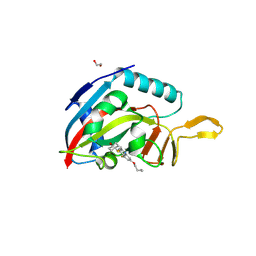 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN012759 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylmethoxy)-5-fluoro-2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}quinazolin-4(3H)-one, Isoform 1 of Protein mono-ADP-ribosyltransferase PARP14 | | Authors: | Swinger, K.K, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
4XDP
 
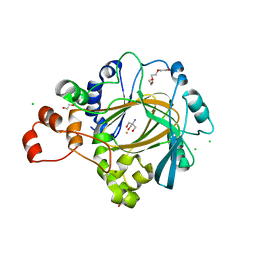 | | Crystal structure of human KDM4C catalytic domain bound to tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A High-Throughput Mass Spectrometry Assay Coupled with Redox Activity Testing Reduces Artifacts and False Positives in Lysine Demethylase Screening.
J Biomol Screen, 20, 2015
|
|
4XDO
 
 | | Crystal structure of human KDM4C catalytic domain with OGA | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Lysine-specific demethylase 4C, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A High-Throughput Mass Spectrometry Assay Coupled with Redox Activity Testing Reduces Artifacts and False Positives in Lysine Demethylase Screening.
J Biomol Screen, 20, 2015
|
|
4Y30
 
 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
4Y2H
 
 | |
1P51
 
 | | Anabaena HU-DNA cocrystal structure (AHU6) | | Descriptor: | 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*T)-3', DNA-binding protein HU | | Authors: | Swinger, K.K, Lemberg, K.M, Zhang, Y, Rice, P.A. | | Deposit date: | 2003-04-24 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexible DNA bending in HU-DNA cocrystal structures
Embo J., 22, 2003
|
|
1P78
 
 | | Anabaena HU-DNA cocrystal structure (AHU2) | | Descriptor: | 5'-D(*TP*GP*CP*AP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3', DNA-binding protein HU | | Authors: | Swinger, K.S, Lemberg, K.M, Zhang, Y, Rice, P.A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Flexible DNA bending in HU-DNA cocrystal structures
Embo J., 22, 2003
|
|
1P71
 
 | | Anabaena HU-DNA corcrystal structure (TR3) | | Descriptor: | 5'-D(*TP*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3', DNA-binding protein HU | | Authors: | Swinger, K.S, Lemberg, K.M, Zhang, Y, Rice, P.A. | | Deposit date: | 2003-04-30 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flexible DNA bending in HU-DNA cocrystal structures
Embo J., 22, 2003
|
|
6CEN
 
 | | Crystal Structure of WHSC1L1 in Complex with Inhibitor PEP21 | | Descriptor: | ACE-GLY-VAL-NLE-ARG-ILE-NH2, Histone-lysine N-methyltransferase NSD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Boriack-Sjodin, P.A, Swinger, K, Farrow, N.A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification of a peptide inhibitor for the histone methyltransferase WHSC1.
PLoS ONE, 13, 2018
|
|
4IQ6
 
 | | Gsk-3beta with inhibitor 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine | | Descriptor: | 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine, Glycogen synthase kinase-3 beta | | Authors: | Tong, Y, Stewart, K.D, Florjancic, A.S, Harlan, J.E, Merta, P.J, Przytulinska, M, Soni, N, Swinger, K.S, Zhu, H, Johnson, E.F, Shoemaker, A.R, Penning, T.D. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Azaindole-Based Inhibitors of Cdc7 Kinase: Impact of the Pre-DFG Residue, Val 195.
ACS Med Chem Lett, 4, 2013
|
|
2A2E
 
 | | Crystal structure of the RNA subunit of Ribonuclease P. Bacterial A-type. | | Descriptor: | OSMIUM ION, RNA subunit of RNase P | | Authors: | Torres-Larios, A, Swinger, K.K, Krasilnikov, A.S, Pan, T, Mondragon, A. | | Deposit date: | 2005-06-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the RNA component of bacterial ribonuclease P.
Nature, 437, 2005
|
|
7L9Y
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN012042 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopentylamino)-5-fluoro-2-{[(piperidin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, ... | | Authors: | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
7LUN
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN011980 | | Descriptor: | 7-(cyclopentylamino)-5-fluoro-2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
5DAF
 
 | | Crystal Structure of Human KEAP1 BTB Domain in Complex with Small Molecule TX64063 | | Descriptor: | (5aS,6S,9aS)-7-hydroxy-2,6,9a-trimethyl-3-(pyridin-3-yl)-4,5,5a,6,9,9a-hexahydro-2H-benzo[g]indazole-8-carbonitrile, Kelch-like ECH-associated protein 1 | | Authors: | Huerta, C, Jiang, X, Trevino, I, Bender, C.F, Swinger, K.K, Stoll, V.S, Ferguson, D.A, Thomas, P.J, Probst, B, Dulubova, I, Visnick, M, Wigley, W.C. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding.
Biochim.Biophys.Acta, 1860, 2016
|
|
5DAD
 
 | | Crystal Structure of Human KEAP1 BTB Domain in Complex with Small Molecule TX64014 | | Descriptor: | (6aS,7S,10aS)-8-hydroxy-4-methoxy-2,7,10a-trimethyl-5,6,6a,7,10,10a-hexahydrobenzo[h]quinazoline-9-carbonitrile, Kelch-like ECH-associated protein 1 | | Authors: | Huerta, C, Jiang, X, Trevino, I, Bender, C.F, Swinger, K.K, Stoll, V.S, Ferguson, D.A, Thomas, P.J, Probst, B, Dulubova, I, Visnick, M, Wigley, W.C. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding.
Biochim.Biophys.Acta, 1860, 2016
|
|
4OQ5
 
 | | Crystal Structure of Human MCL-1 Bound to Inhibitor 4-(4-methylnaphthalen-1-yl)-2-{[(4-phenoxyphenyl)sulfonyl]amino}benzoic acid | | Descriptor: | 4-(4-methylnaphthalen-1-yl)-2-{[(4-phenoxyphenyl)sulfonyl]amino}benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Petros, A.M, Swann, S.L, Song, D, Swinger, K, Park, C, Zhang, H, Wendt, M.D, Kunzer, A.R, Souers, A.J, Sun, C. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Fragment-based discovery of potent inhibitors of the anti-apoptotic MCL-1 protein.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3Q1R
 
 | | Crystal structure of a bacterial RNase P holoenzyme in complex with TRNA and in the presence of 5' leader | | Descriptor: | MAGNESIUM ION, RNase P RNA, Ribonuclease P protein component, ... | | Authors: | Reiter, N.J, Ostermanm, A, Torres-Larios, A, Swinger, K.K, Pan, T, Mondragon, A. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.21 Å) | | Cite: | Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with tRNA.
Nature, 468, 2010
|
|
3Q1Q
 
 | | Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with tRNA | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, RNase P RNA, ... | | Authors: | Reiter, N.J, Osterman, A, Torres-Larios, A, Swinger, K.K, Pan, T, Mondragon, A. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with tRNA.
Nature, 468, 2010
|
|
4OQ6
 
 | | Crystal Structure of Human MCL-1 Bound to Inhibitor 4-hydroxy-4'-propylbiphenyl-3-carboxylic acid | | Descriptor: | 4-hydroxy-4'-propylbiphenyl-3-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Petros, A.M, Swann, S.L, Song, D, Swinger, K, Park, C, Zhang, H, Wendt, M.D, Kunzer, A.R, Souers, A.J, Sun, C. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of potent inhibitors of the anti-apoptotic MCL-1 protein.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
