6XPG
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography | | Descriptor: | Lysophospholipase L1 | | Authors: | Kim, Y, Sherrell, D.A, Owen, R, Axford, D, Ebrahim, A, Johnson, J, Welk, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography
To Be Published
|
|
4FUM
 
 | |
7RSK
 
 | | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus | | Descriptor: | Glycosyl hydrolase family 2, sugar binding domain protein | | Authors: | Kim, Y, Nocek, B, Endres, M, Joachimiak, G, Johnson, J, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-08-11 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus
To Be Published
|
|
6BO3
 
 | | Structure Determination of A223, a turret protein in Sulfolobus turreted icosahedral virus, using an iterative hybrid approach | | Descriptor: | Uncharacterized protein | | Authors: | Sendamarai, A.K, Veesler, D, Fu, C.Y, Marceau, C, Larson, E.T, Johnson, J.E, Lawrence, C.M. | | Deposit date: | 2017-11-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A223, A STRUCTURAL PROTEIN FROM SULFOLOBUS TURRETED ICOSAHEDRAL VIRUS (STIV)
To Be Published
|
|
6F57
 
 | |
3CZD
 
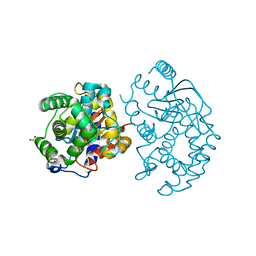 | | Crystal structure of human glutaminase in complex with L-glutamate | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutaminase kidney isoform, ... | | Authors: | Karlberg, T, Welin, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wikstrom, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ZGA
 
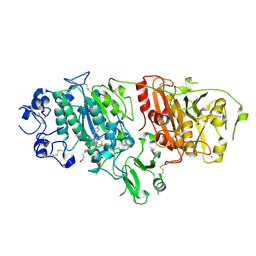 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (11aS)-6-(4-fluorobenzyl)-5,6,11,11a-tetrahydro-1H-imidazo[1',5':1,6]pyrido[3,4-b]indole-1,3(2H)-dione, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
3EC8
 
 | | The crystal structure of the RA domain of FLJ10324 (RADIL) | | Descriptor: | CHLORIDE ION, GLYCEROL, LEAD (II) ION, ... | | Authors: | Wisniewska, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the RA domain of FLJ10324 (RADIL)
to be published
|
|
4ZG7
 
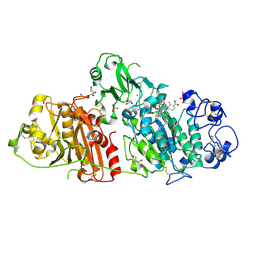 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-({6-chloro-7-fluoro-2-methyl-1-[2-oxo-2-(spiro[cyclopropane-1,3'-indol]-1'(2'H)-yl)ethyl]-1H-indol-3-yl}sulfanyl)-2-fluorobenzoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
8EIH
 
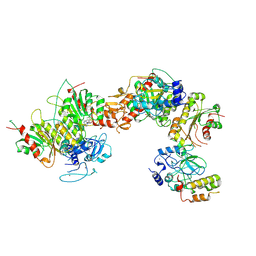 | | Cryo-EM structure of human DNMT3B homo-tetramer (form I) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIK
 
 | | Cryo-EM structure of human DNMT3B homo-hexamer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIJ
 
 | | Cryo-EM structure of human DNMT3B homo-trimer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EII
 
 | | Cryo-EM structure of human DNMT3B homo-tetramer (form II) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
4ZG6
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 4-{(Z)-2-[6-chloro-1-(4-fluorobenzyl)-1H-indol-3-yl]-1-cyanoethenyl}benzoic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
7DAG
 
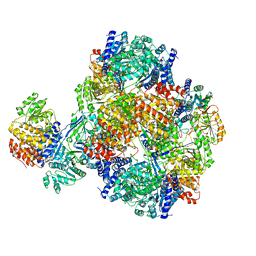 | | Vibrio cholera aldehyde-alcohol dehrogenase | | Descriptor: | Aldehyde-alcohol dehydrogenase | | Authors: | Cho, S, Cho, C, Song, J, Kim, G. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | Cryo-EM structure of Vibrio cholerae aldehyde-alcohol dehydrogenase spirosomes.
Biochem.Biophys.Res.Commun., 536, 2020
|
|
1F8V
 
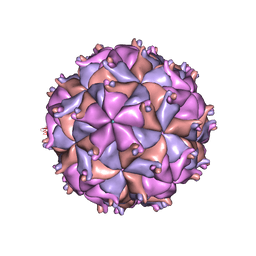 | | THE STRUCTURE OF PARIACOTO VIRUS REVEALS A DODECAHEDRAL CAGE OF DUPLEX RNA | | Descriptor: | CALCIUM ION, MATURE CAPSID PROTEIN BETA, MATURE CAPSID PROTEIN GAMMA, ... | | Authors: | Tang, L, Johnson, K.N, Ball, L.A, Lin, T, Yeager, M, Johnson, J.E. | | Deposit date: | 2000-07-05 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA.
Nat.Struct.Biol., 8, 2001
|
|
6GCX
 
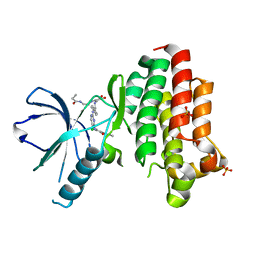 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | Descriptor: | 2-[[2-[[4-[[[3,4-bis(oxidanylidene)-2-[2-(propanoylamino)ethylamino]cyclobuten-1-yl]amino]methyl]phenyl]amino]-5-chloranyl-pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1, SULFATE ION | | Authors: | Yen-Pon, E, Li, B, Acebron-Garcia de Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
6VJV
 
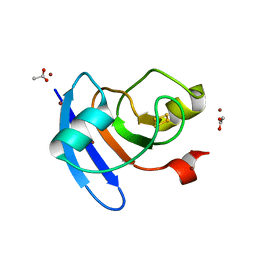 | | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | | Descriptor: | ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Olmos Jr, J.L, Campbell, I.J, Miller, M.D, Xu, W, Kahanda, D, Atkinson, J.T, Sparks, N, Bennett, G.N, Silberg, J.J, Phillips Jr, G.N. | | Deposit date: | 2020-01-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases.
J.Biol.Chem., 295, 2020
|
|
7SI4
 
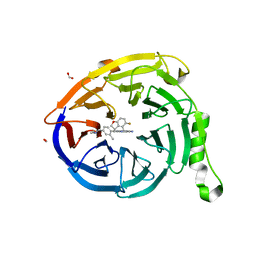 | | CRYSTAL STRUCTURE OF EED WITH MRTX-2219 | | Descriptor: | (4S)-8-{4-[(dimethylamino)methyl]-2-methylphenyl}-5-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}imidazo[1,2-c]pyrimidine-2-carbonitrile, FORMIC ACID, Polycomb protein EED | | Authors: | Gunn, R.J, Burns, A.C, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF EED WITH MRTX-2219
TO BE PUBLISHED
|
|
6GCR
 
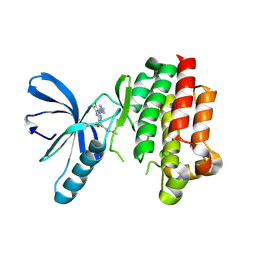 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | Descriptor: | 2-[[2-[[4-[[[3,4-bis(oxidanylidene)-2-[2-(propanoylamino)ethylamino]cyclobuten-1-yl]amino]methyl]phenyl]amino]-5-chloranyl-pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1 | | Authors: | Yen-Pon, E, Li, B, Acebron-Garcia de Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
2WSA
 
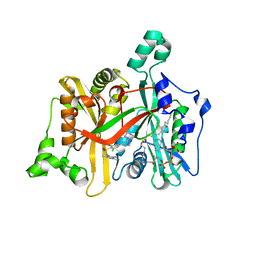 | | Crystal Structure of Leishmania major N-myristoyltransferase (NMT) with bound myristoyl-CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | N-Myristoyltransferase Inhibitors as New Leads to Treat Sleeping Sickness.
Nature, 464, 2010
|
|
7SZL
 
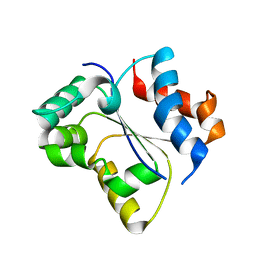 | | Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1R10 (IL-1RAPL2) | | Descriptor: | X-linked interleukin-1 receptor accessory protein-like 2 | | Authors: | Nimma, S, Gu, W, Manik, M.K, Ve, T, Nanson, J.D, Kobe, B. | | Deposit date: | 2021-11-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Toll/interleukin-1 receptor (TIR) domain of IL-1R10 provides structural insights into TIR domain signalling.
Febs Lett., 596, 2022
|
|
1NY7
 
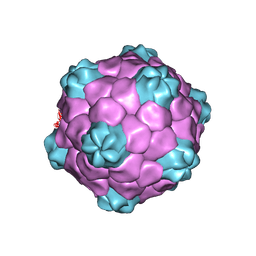 | | COWPEA MOSAIC VIRUS (CPMV) | | Descriptor: | COWPEA MOSAIC VIRUS, LARGE (L) SUBUNIT, SMALL (S) SUBUNIT | | Authors: | Lin, T, Chen, Z, Usha, R, Stauffacher, C.V, Dai, J.-B, Schmidt, T, Johnson, J.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Refined Crystal Structure of Cowpea Mosaic Virus at 2.8A Resolution
Virology, 265, 1999
|
|
8FAK
 
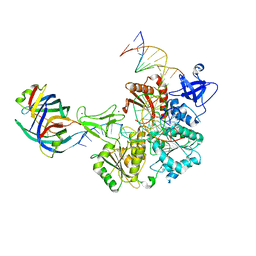 | | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | | Descriptor: | DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Duckworth, A.T, Ducos, P.L, McMillan, S.D, Satyshur, K.A, Blumenthal, K.H, Deorio, H.R, Larson, J.A, Sandler, S.J, Grant, T, Keck, J.L. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli.
Nat Commun, 14, 2023
|
|
3F0W
 
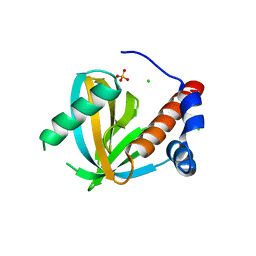 | | Human NUMB-like protein, phosphotyrosine interaction domain | | Descriptor: | CHLORIDE ION, Numb-like protein, SULFATE ION | | Authors: | Lehtio, L, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, D Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human NUMB-like protein, phosphotyrosine interaction domain
To be Published
|
|
