6QBB
 
 | |
6QSY
 
 | |
6QMU
 
 | |
6QW4
 
 | |
6GR0
 
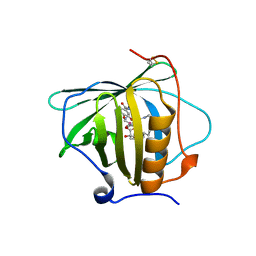 | | Petrobactin-binding engineered lipocalin in complex with gallium-petrobactin | | Descriptor: | 4-[4-[3-[[3,4-bis(oxidanyl)phenyl]carbonylamino]propylamino]butylamino]-2-[2-[4-[3-[[3,4-bis(oxidanyl)phenyl]carbonylamino]propylamino]butylamino]-2-oxidanylidene-ethyl]-2-oxidanyl-4-oxidanylidene-butanoic acid, GALLIUM (III) ION, Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A. | | Deposit date: | 2018-06-08 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reprogramming Human Siderocalin To Neutralize Petrobactin, the Essential Iron Scavenger of Anthrax Bacillus.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GQZ
 
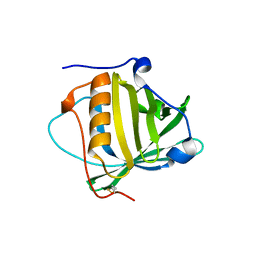 | | Petrobactin-binding engineered lipocalin without ligand | | Descriptor: | Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A. | | Deposit date: | 2018-06-08 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reprogramming Human Siderocalin To Neutralize Petrobactin, the Essential Iron Scavenger of Anthrax Bacillus.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6ZVF
 
 | |
6Z2C
 
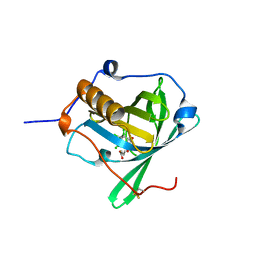 | | Engineered lipocalin C3A5 in complex with a transition state analog | | Descriptor: | 1,7,8,9,10,10-hexachloro-4-carboxypentyl-4-aza-tricyclo[5.2.1.0(2,6)]dec-8-ene-3,5-dione, Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A. | | Deposit date: | 2020-05-15 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an engineered lipocalin that catalyzes a Diels-Alder reaction
To be published
|
|
6Z6Z
 
 | |
3P7G
 
 | |
3P7F
 
 | | Structure of the human Langerin carbohydrate recognition domain | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION | | Authors: | Skerra, A, Schiefner, A. | | Deposit date: | 2010-10-12 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The carbohydrate recognition domain of Langerin reveals high structural similarity with the one of DC-SIGN but an additional, calcium-independent sugar-binding site.
Mol.Immunol., 45, 2008
|
|
3P7H
 
 | |
6SOK
 
 | |
6SOS
 
 | |
6TIP
 
 | |
5NKN
 
 | | Crystal structure of an Anticalin-colchicine complex | | Descriptor: | N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide, Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A, Barkovskiy, M. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An engineered lipocalin that tightly complexes the plant poison colchicine for use as antidote and in bioanalytical applications.
Biol. Chem., 400, 2019
|
|
5NBW
 
 | |
5MHH
 
 | |
8B50
 
 | |
5BPK
 
 | | Varying binding modes of inhibitors and structural differences in the binding pockets of different gamma-glutamyltranspeptidases | | Descriptor: | (2S)-amino[(5S)-4,5-dihydro-1,2-oxazol-5-yl]acetic acid, 1,2-ETHANEDIOL, Gamma-glutamyltranspeptidase (Ggt) | | Authors: | Bolz, C, Bach, N.C, Meyer, H, Mueller, G, Dawidowski, M, Popowicz, G, Sieber, S.A, Skerra, A, Gerhard, M. | | Deposit date: | 2015-05-28 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Varying binding modes of inhibitors and structural differences in the binding pockets of different gamma-glutamyltranspeptidases
To Be Published
|
|
7P85
 
 | |
6I9Q
 
 | | Structure of the mouse CD98 heavy chain ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, CHLORIDE ION | | Authors: | Schiefner, A, Deuschle, F.-C, Skerra, A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural differences between the ectodomains of murine and human CD98hc.
Proteins, 87, 2019
|
|
7O31
 
 | | Crystal structure of the anti-PAS Fab 1.2 in complex with its epitope peptide and the anti-Kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, PAS#1 epitope peptide, anti-Kappa VHH domain, ... | | Authors: | Schilz, J, Schiefner, A, Skerra, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
7O2Z
 
 | | Crystal structure of the anti-PAS Fab 2.2 in complex with its epitope peptide | | Descriptor: | CHLORIDE ION, P/A#1 epitope peptide, anti-PAS Fab 2.2 chimeric heavy chain, ... | | Authors: | Schilz, J, Schiefner, A, Skerra, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
7O30
 
 | |
