8TRS
 
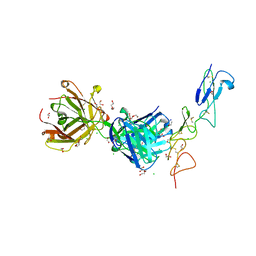 | | Structure of the EphA2 CRD bound to FabS1CE_C1, trigonal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
2K8Y
 
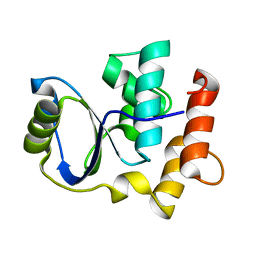 | | Solution NMR Structure of Cgi121 from Methanococcus jannaschii. Northeast Structural Genomics Consortium Target MJ0187 | | Descriptor: | Uncharacterized protein MJ0187 | | Authors: | Rumpel, S, Fares, C, Neculai, D, Arrowsmith, C, Montelione, G.T, Sicheri, F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
2JQZ
 
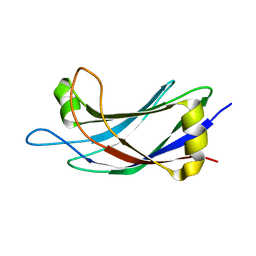 | | Solution Structure of the C2 domain of human Smurf2 | | Descriptor: | E3 ubiquitin-protein ligase SMURF2 | | Authors: | Wiesner, S, Ogunjimi, A.A, Wang, H, Rotin, D, Sicheri, F, Wrana, J.L, Forman-Kay, J.D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of the HECT-Type Ubiquitin Ligase Smurf2 through Its C2 Domain
Cell(Cambridge,Mass.), 130, 2007
|
|
2PBC
 
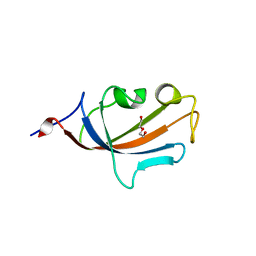 | | FK506-binding protein 2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FK506-binding protein 2 | | Authors: | Walker, J.R, Neculai, D, Davis, T, Butler-Cole, C, Sicheri, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of FK506-Binding Protein 2
To be Published
|
|
2HEL
 
 | |
2KIS
 
 | | Solution structure of CA150 FF1 domain and FF1-FF2 interdomain linker | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Murphy, J.M, Hansen, D, Wiesner, S, Muhandiram, D, Borg, M, Smith, M.J, Sicheri, F, Kay, L.E, Forman-Kay, J.D, Pawson, T. | | Deposit date: | 2009-05-08 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural studies of FF domains of the transcription factor CA150 provide insights into the organization of FF domain tandem arrays.
J.Mol.Biol., 393, 2009
|
|
4PDS
 
 | | Crystal structure of Rad53 kinase domain and SCD2 in complex with AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase RAD53 | | Authors: | Ho, C.S, Wybenga-Groot, L.E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2014-04-21 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of Rad53 kinase activation by dimerization and activation segment exchange.
Cell Signal., 26, 2014
|
|
4NKG
 
 | | Crystal structure of SspH1 LRR domain in complex PKN1 HR1b domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1, HEXANE-1,6-DIOL, Serine/threonine-protein kinase N1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
6CRN
 
 | | Structure of the USP15 deubiquitinase domain in complex with a high-affinity first-generation Ubv | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 15, Ubiquitin variant 15.2, ZINC ION | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-19 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
6BVA
 
 | |
6BYH
 
 | |
6DJ9
 
 | | Structure of the USP15 DUSP domain in complex with a high-affinity Ubiquitin Variant (UbV) | | Descriptor: | Ubiquitin Variant UbV 15.D, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-05-24 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
4PDP
 
 | |
6C16
 
 | |
6CPM
 
 | | Structure of the USP15 deubiquitinase domain in complex with a third-generation inhibitory Ubv | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-13 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
4O1O
 
 | | Crystal Structure of RNase L in complex with 2-5A | | Descriptor: | Ribonuclease L, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
4NKH
 
 | | Crystal structure of SspH1 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
4PL3
 
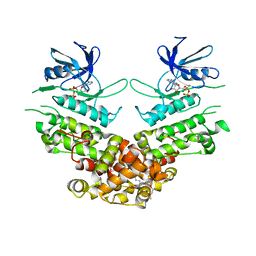 | | Crystal structure of murine IRE1 in complex with MKC9989 inhibitor | | Descriptor: | 7-hydroxy-6-methoxy-3-[2-(2-methoxyethoxy)ethyl]-4,8-dimethyl-2H-chromen-2-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
6D4P
 
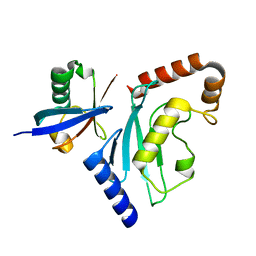 | | Ube2D1 in complex with ubiquitin variant Ubv.D1.1 | | Descriptor: | Ubiquitin Variant Ubv.D1.1, Ubiquitin-conjugating enzyme E2 D1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D68
 
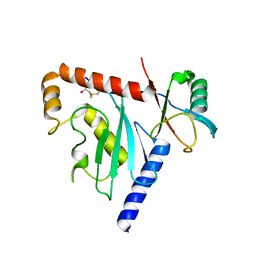 | | Ube2G1 in complex with ubiquitin variant Ubv.G1.1 | | Descriptor: | Ubiquitin-conjugating enzyme E2 G1, Ubv.G1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-20 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
4PL4
 
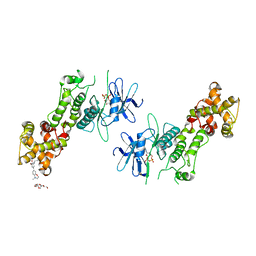 | | Crystal structure of murine IRE1 in complex with OICR464 inhibitor | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-methoxy-6-methyl-4-(4-methyl-3,4-dihydro-2H-1,4-benzoxazin-7-yl)phenol, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
6DGF
 
 | | Ubiquitin Variant bound to USP2 | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 2, ... | | Authors: | Manczyk, N, Sicheri, F. | | Deposit date: | 2018-05-17 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Yeast Two-Hybrid Analysis for Ubiquitin Variant Inhibitors of Human Deubiquitinases.
J. Mol. Biol., 431, 2019
|
|
6D6I
 
 | | Ube2V1 in complex with ubiquitin variant Ubv.V1.1 and Ube2N/Ubc13 | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1, Ubv.V1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Keszei, A, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6CAD
 
 | | Crystal structure of RAF kinase domain bound to the inhibitor 2a | | Descriptor: | 1-(propan-2-yl)-3-({3-[3-(trifluoromethyl)phenyl]isoquinolin-8-yl}ethynyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Liu, X, Chen, Y.-C, Prakash, G.K.S, Zhang, C, SIcheri, F. | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effects of rigidity on the selectivity of protein kinase inhibitors.
Eur J Med Chem, 146, 2018
|
|
6BXI
 
 | | X-ray crystal structure of NDR1 kinase domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase 38 | | Authors: | Xiong, S, Sicheri, F. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Auto-Inhibition of the NDR1 Kinase Domain by an Atypically Long Activation Segment.
Structure, 26, 2018
|
|
