8IPA
 
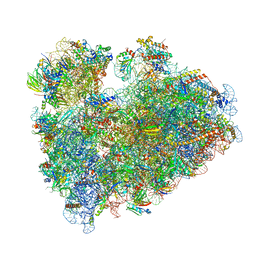 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently with cycloheximide | | Descriptor: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
1Q5D
 
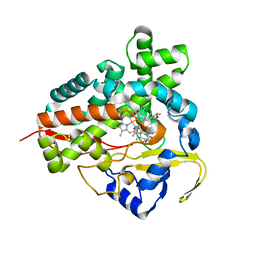 | | Epothilone B-bound Cytochrome P450epoK | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
5TYR
 
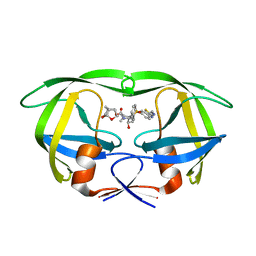 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
1Q5E
 
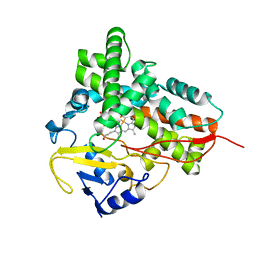 | | Substrate-free Cytochrome P450epoK | | Descriptor: | P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
4H04
 
 | | Lacto-N-biosidase from Bifidobacterium bifidum | | Descriptor: | Lacto-N-biosidase, SULFATE ION, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ito, T, Katayama, T, Wada, J, Suzuki, R, Ashida, H, Wakagi, T, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2012-09-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
4EN8
 
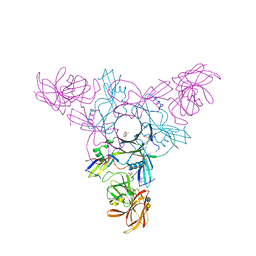 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN9
 
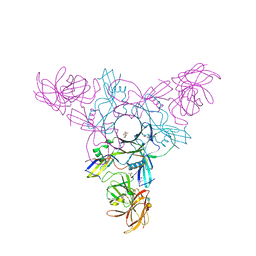 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
1IUG
 
 | | The crystal structure of aspartate aminotransferase which belongs to subgroup IV from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, putative aspartate aminotransferase | | Authors: | Katsura, Y, Shirouzu, M, Yamaguchi, H, Ishitani, R, Nureki, O, Kuramitsu, S, Hayashi, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-04 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative aspartate aminotransferase belonging to subgroup IV.
Proteins, 55, 2004
|
|
5B13
 
 | | Crystal structure of phycoerythrin | | Descriptor: | PHYCOCYANOBILIN, PHYCOUROBILIN, Phycoerythrin alpha subunit, ... | | Authors: | Tanaka, Y, Gai, Z, Kishimura, H. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structural properties of phycoerythrin from dulse palmaria palmata
J FOOD BIOCHEM., 2016
|
|
6NU4
 
 | | Solution structure of the Arabidopsis thaliana RALF8 peptide | | Descriptor: | Protein RALF-like 8 | | Authors: | Lee, W, Markley, J.L, Frederick, R.O, Miyoshi, H, Tonelli, M, Cornilescu, G, Cornilescu, C, Sussman, M.R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Function and solution structure of the Arabidopsis thaliana RALF8 peptide.
Protein Sci., 28, 2019
|
|
4EN7
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
8X5V
 
 | | BlCas9-sgRNA-target DNA complex | | Descriptor: | 1,2-ETHANEDIOL, BlCas9, CHLORIDE ION, ... | | Authors: | Nakane, T, Nakagawa, R, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2023-11-19 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and engineering of Brevibacillus laterosporus Cas9.
Commun Biol, 7, 2024
|
|
4EN6
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
4L3O
 
 | | Crystal Structure of SIRT2 in complex with the macrocyclic peptide S2iL5 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Yamagata, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-06-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Structural Basis for Potent Inhibition of SIRT2 Deacetylase by a Macrocyclic Peptide Inducing Dynamic Structural Change
Structure, 22, 2013
|
|
1J0S
 
 | | Solution structure of the human interleukin-18 | | Descriptor: | Interleukin-18 | | Authors: | Kato, Z, Jee, J, Shikano, H, Mishima, M, Ohki, I, Yoneda, T, Hara, T, Torigoe, K, Kondo, N, Shirakawa, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structure and binding mode of interleukin-18
Nat.Struct.Biol., 10, 2003
|
|
5GIJ
 
 | | Crystal structure of TDR-TDIF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat receptor-like protein kinase TDR, ... | | Authors: | Morita, J, Kato, K, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide
Nat Commun, 7, 2016
|
|
6JPL
 
 | | The X-ray structure of yeast tRNA methyltransferase Trm7-Trm734 in complex with S-adenosyl-L-methionine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Hirata, A, Okada, K, Yoshii, K, Shiraisi, H, Saijo, S, Yonezawa, K, Shimizu, N, Hori, H. | | Deposit date: | 2019-03-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
4P1G
 
 | | Crystal structure of the Bateman domain of murine magnesium transporter CNNM2 bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Abascal-Palacios, G, Diercks, T, Oyenarte, I, Ereno-Orbea, J, Encinar, J.A, Spiwok, V, Terashima, H, Accardi, A, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To Be Published
|
|
5HRT
 
 | | Crystal structure of mouse autotaxin in complex with a DNA aptamer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-01-24 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis for specific inhibition of Autotaxin by a DNA aptamer
Nat.Struct.Mol.Biol., 23, 2016
|
|
7V6B
 
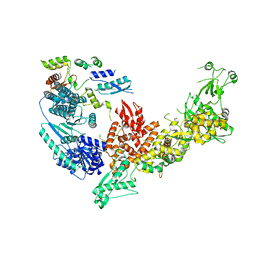 | | Structure of the Dicer-2-R2D2 heterodimer | | Descriptor: | Dicer-2, isoform A, R2D2 | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
7V6C
 
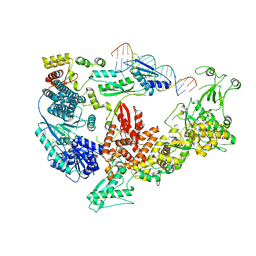 | | Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex | | Descriptor: | Dicer-2, isoform A, R2D2, ... | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
4P1O
 
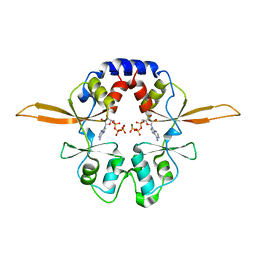 | | Crystal structure of the Bateman domain of murine magnesium transporter CNNM2 bound to ATP-Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Abascal-Palacios, G, Diercks, T, Oyenarte, I, Ereno-Orbea, J, Encinar, J.A, Spiwok, V, Terashima, H, Accardi, A, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To Be Published
|
|
3VU7
 
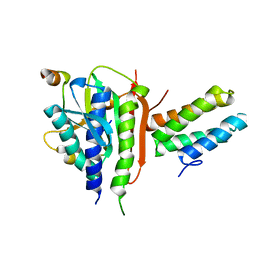 | | Crystal structure of REV1-REV7-REV3 ternary complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Kikuchi, S, Hara, K, Shimizu, T, Sato, M, Hashimoto, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of recruitment of DNA polymerase [zeta] by interaction between REV1 and REV7 proteins
J.Biol.Chem., 287, 2012
|
|
5GUH
 
 | | Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA | | Descriptor: | MAGNESIUM ION, PIWI, RNA (28-MER) | | Authors: | Matsumoto, N, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA
Cell, 167, 2016
|
|
