8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-15 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
5LIG
 
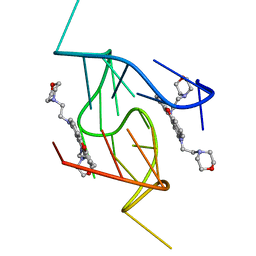 | | G-Quadruplex formed at the 5'-end of NHEIII_1 Element in human c-MYC promoter bound to triangulenium based fluorescence probe DAOTA-M2 | | 分子名称: | 8,12-bis(2-morpholinoethyl)-8H-benzo[ij]xantheno[1,9,8-cdef][2,7]naphthyridin-12-iumhexafluorophosphate, DNA (5'-D(*TP*AP*GP*GP*GP*AP*GP*GP*GP*TP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3') | | 著者 | Kotar, A, Wang, B, Shivalingam, A, Gonzalez-Garcia, J, Vilar, R, Plavec, J. | | 登録日 | 2016-07-14 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of a Triangulenium-Based Long-Lived Fluorescence Probe Bound to a G-Quadruplex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | 著者 | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-08 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
5NFF
 
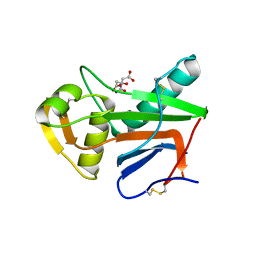 | | Crystal structure of GP1 receptor binding domain from Morogoro virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | 著者 | Israeli, H, Cohen-Dvashi, H, Shulman, A, Shimon, A, Diskin, R. | | 登録日 | 2017-03-14 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.615 Å) | | 主引用文献 | Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses.
PLoS Pathog., 13, 2017
|
|
8E40
 
 | | Full-length APOBEC3G in complex with HIV-1 Vif, CBF-beta, and fork RNA | | 分子名称: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, RNA, ... | | 著者 | Ito, F, Alvarez-Cabrera, A.L, Liu, S, Yang, H, Shiriaeva, A, Zhou, Z.H, Chen, X.S. | | 登録日 | 2022-08-17 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | Structural basis for HIV-1 antagonism of host APOBEC3G via Cullin E3 ligase.
Sci Adv, 9, 2023
|
|
4U9P
 
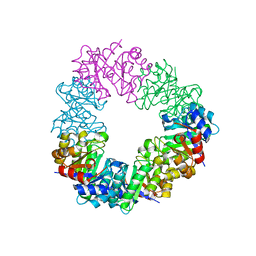 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii | | 分子名称: | GLYCEROL, UPF0264 protein MJ1099 | | 著者 | Bobik, T.A, Morales, E, Shin, A, Cascio, D, Sawaya, M.R, Arbing, M, Rasche, M.E. | | 登録日 | 2014-08-06 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
6VCQ
 
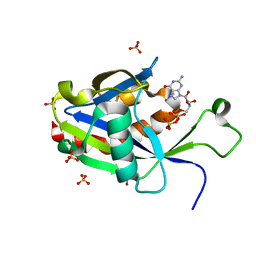 | | Crystal structure of E.coli RppH in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, RNA pyrophosphohydrolase, SULFATE ION | | 著者 | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | 登録日 | 2019-12-21 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
3ALR
 
 | | Crystal structure of Nanos | | 分子名称: | Nanos protein, ZINC ION | | 著者 | Hashimoto, H, Hara, K, Hishiki, A, Kawaguchi, S, Shichijo, N, Nakamura, K, Unzai, S, Tamaru, Y, Shimizu, T, Sato, M. | | 登録日 | 2010-08-06 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of zinc-finger domain of Nanos and its functional implications
Embo Rep., 11, 2010
|
|
6VCN
 
 | | Crystal structure of E.coli RppH in complex with ppcpG | | 分子名称: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, RNA pyrophosphohydrolase, SULFATE ION | | 著者 | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | 登録日 | 2019-12-21 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
8QF2
 
 | | Beta-L-Arabinofurano-cyclitol Aziridines are Cysteine-directed Broad-spectrum Inhibitors and Activity-based Probes for Retaining Beta-L-arabinofuranosidases | | 分子名称: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Borlandelli, V, Offen, W.A, Moroz, O, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | 登録日 | 2023-09-02 | | 公開日 | 2023-12-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
8QF8
 
 | | GH146 beta-L-arabinofuranosidase from Bacteroides thetaioatomicron in complex with beta-l-arabinofurano cyclophellitol aziridine | | 分子名称: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, (1~{S},2~{S},3~{S},4~{S},5~{S})-4-(hydroxymethyl)-6-azabicyclo[3.1.0]hexane-2,3-diol, Glycosyl hydrolase, ... | | 著者 | Borlandelli, V, Offen, W, Moroz, O.V, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | 登録日 | 2023-09-04 | | 公開日 | 2023-12-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
8SF9
 
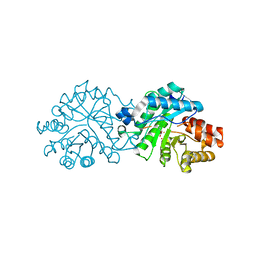 | | Crystal structure of the engineered SsoPox variant IG7 - Alternative state | | 分子名称: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION | | 著者 | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | 登録日 | 2023-04-10 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
6VCR
 
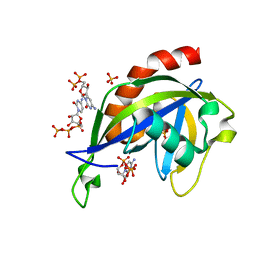 | | Crystal structure of E.coli RppH in complex with CTP | | 分子名称: | CYTIDINE-5'-TRIPHOSPHATE, PYROPHOSPHATE, RNA pyrophosphohydrolase, ... | | 著者 | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | 登録日 | 2019-12-21 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
5XDX
 
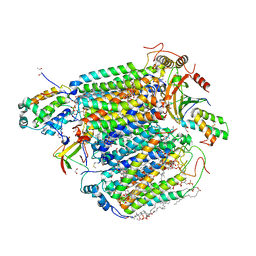 | | Bovine heart cytochrome c oxidase in the reduced state with pH 7.3 at 1.99 angstrom resolution | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | 登録日 | 2017-03-30 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure of bovine cytochrome c oxidase in the ligand-free reduced state at neutral pH.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5XDQ
 
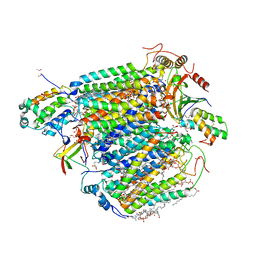 | | Bovine heart cytochrome c oxidase in the fully oxidized state with pH 7.3 at 1.77 angstrom resolution | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | 登録日 | 2017-03-29 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structure of bovine cytochrome c oxidase crystallized at a neutral pH using a fluorinated detergent.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1IPB
 
 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GPPPA | | 分子名称: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | 著者 | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | 登録日 | 2001-05-08 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
1IST
 
 | |
1IPC
 
 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GTP | | 分子名称: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | 著者 | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | 登録日 | 2001-05-08 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
8SFD
 
 | | Crystal structure of the engineered SsoPox variant IVB10 | | 分子名称: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | 著者 | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | 登録日 | 2023-04-10 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFK
 
 | | Crystal structure of the engineered SsoPox variant IVE2 | | 分子名称: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | 著者 | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | 登録日 | 2023-04-11 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFB
 
 | | Crystal structure of the engineered SsoPox variant IVA4 | | 分子名称: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | 著者 | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | 登録日 | 2023-04-10 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFC
 
 | | Crystal structure of the engineered SsoPox variant IVA4 in alternate state | | 分子名称: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | 著者 | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | 登録日 | 2023-04-10 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
6VCK
 
 | | Crystal structure of E.coli RppH-DapF in complex with GDP, Mg2+ and F- | | 分子名称: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | 著者 | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | 登録日 | 2019-12-21 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCP
 
 | | Crystal structure of E.coli RppH in complex with UTP | | 分子名称: | RNA pyrophosphohydrolase, URIDINE 5'-TRIPHOSPHATE | | 著者 | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | 登録日 | 2019-12-21 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
8DZD
 
 | |
