6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | Descriptor: | DNA repair protein RAD5, ZINC ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
5NPQ
 
 | | Human N-myristoyltransferase 1 (MNT1) with Myristoyl-CoA analogue X10 bound | | Descriptor: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, MAGNESIUM ION, ... | | Authors: | Shen, M, Perez-Dorado, I, Fedoryshchak, R, Tate, E.W. | | Deposit date: | 2017-04-18 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Human N-myristoyltransferase 1 (MNT1) with Myristoyl-CoA analogue X10 bound.
To be published
|
|
4ZLK
 
 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | Authors: | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
6L8O
 
 | | Crystal structure of the K. lactis Rad5 (Hg-derivative) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA repair protein RAD5, MERCURY (II) ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
7EIM
 
 | |
7EJT
 
 | |
7EJP
 
 | |
7EKX
 
 | |
7EKW
 
 | |
7EKU
 
 | |
4JBH
 
 | | 2.2A resolution structure of cobalt and zinc bound thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
4JBI
 
 | | 2.35A resolution structure of NADPH bound thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
4JBG
 
 | | 1.75A resolution structure of a thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
3SRK
 
 | | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, POTASSIUM ION, Pyruvate kinase, ... | | Authors: | Morgan, H.P, Walsh, M, Blackburn, E.A, Wear, M.A, Boxer, M, Shen, M, McNae, I.W, Michels, P.A.M, Auld, D.S, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase
To be Published
|
|
3IES
 
 | | Firefly luciferase inhibitor complex | | Descriptor: | 5'-O-[(R)-[({3-[5-(2-fluorophenyl)-1,2,4-oxadiazol-3-yl]phenyl}carbonyl)oxy](hydroxy)phosphoryl]adenosine, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IER
 
 | | Firefly luciferase apo structure (P41 form) with PEG 400 bound | | Descriptor: | Luciferin 4-monooxygenase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IEP
 
 | | Firefly luciferase apo structure (P41 form) | | Descriptor: | Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3RIX
 
 | | 1.7A resolution structure of a firefly luciferase-Aspulvinone J inhibitor complex | | Descriptor: | (5Z)-4-hydroxy-3-[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]-5-{[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]methylidene}furan-2(5H)-one, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Lopez, P.C, Auld, D.S, Schultz, P.J, MacArthur, R, Shen, M, Tamayo, G, Inglese, J, Sherman, D.H. | | Deposit date: | 2011-04-14 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Titration-based screening for evaluation of natural product extracts: identification of an aspulvinone family of luciferase inhibitors.
Chem.Biol., 18, 2011
|
|
4H58
 
 | | BRAF in complex with compound 3 | | Descriptor: | CHLORIDE ION, N-(4-{[(2-methoxyethyl)amino]methyl}phenyl)-6-(pyridin-4-yl)quinazolin-2-amine, Serine/threonine-protein kinase B-raf | | Authors: | Vasbinder, M, Aquila, B, Augustin, M, Chueng, T, Cook, D, Drew, L, Fauber, B, Glossop, S, Godin, R, Grondine, M, Hennessy, E, Johannes, J, Lee, S, Lyne, P, Moertl, M, Omer, C, Palakurthi, S, Pontz, T, Read, J, Sha, L, Shen, M, Steinbacher, S, Wang, H, Wu, A, Ye, M, Bagal, B. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and Optimization of a Novel Series of Potent Mutant B-Raf(V600E) Selective Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4E5D
 
 | | 2.2A resolution structure of a firefly luciferase-benzothiazole inhibitor complex | | Descriptor: | 2-(2-fluorophenyl)-6-methoxy-1,3-benzothiazole, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Throne, N, Shen, M, Auld, D.S, Inglese, J. | | Deposit date: | 2012-03-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter.
Chem.Biol., 19, 2012
|
|
6J1K
 
 | | Crystal structure of ribose-5-phosphate isomerase A from Ochrobactrum sp. CSL1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ribose-5-phosphate isomerase A | | Authors: | Xu, X.Q, Mo, X.B, Ju, X, Shen, M, Li, L.Z, Yuan, A.Y. | | Deposit date: | 2018-12-28 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.824 Å) | | Cite: | Crystal structure of ribose-5-phosphate isomerase A from Ochrobactrum sp. CSL1
To be published
|
|
5OWO
 
 | | Human cytoplasmic Dynein N-Terminus dimerization domain at 1.8 Angstrom resolution | | Descriptor: | CALCIUM ION, Cytoplasmic dynein 1 heavy chain 1, GLYCEROL, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
6F1T
 
 | | Cryo-EM structure of two dynein tail domains bound to dynactin and BICDR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
6F3A
 
 | | Cryo-EM structure of a single dynein tail domain bound to dynactin and BICD2N | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
6F1V
 
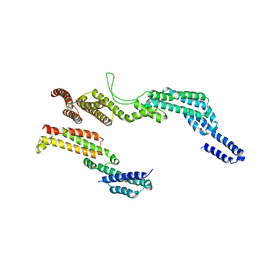 | | C terminal region of the dynein heavy chains in the dynein tail/dynactin/BICDR1 complex | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1 | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
