8VEF
 
 | |
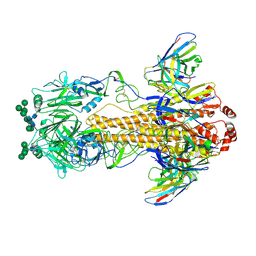
8VEB
 
 | |
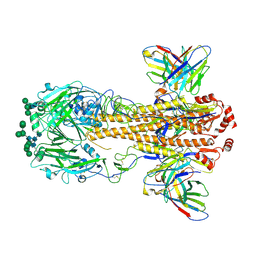
8VEE
 
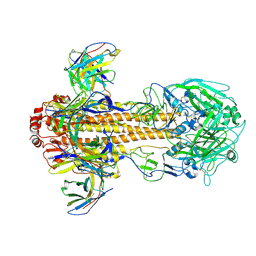 | | Cryo-EM structure of antibody T5-1E08 in complex with H7N9 Influenza Hemagglutinin Trimer (A/Shanghai/2/13) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Cerutti, G, Casner, R.G, Shapiro, L. | | Deposit date: | 2023-12-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Distinct binding modes drive the broad neutralization profile of two persistent influenza hemagglutinin stem-specific antibody lineages.
Structure, 33, 2025
|
|
8DW9
 
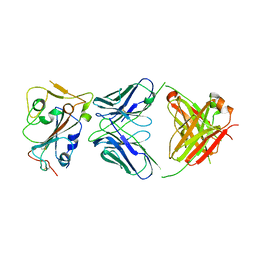 | |
8DXS
 
 | |
8DWA
 
 | |
6U3Z
 
 | | Structure of VD20_5A4 Fab | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dionne, G, Shapiro, L. | | Deposit date: | 2019-08-22 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99002314 Å) | | Cite: | Extensive dissemination and intraclonal maturation of HIV Env vaccine-induced B cell responses.
J.Exp.Med., 217, 2020
|
|
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | Descriptor: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L, Ho, D.D. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
8DUL
 
 | | Cryo-EM Structure of Antibody SKT05 in complex with Western Equine Encephalitis Virus spike (local refinement from VLP particles) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab SKT05 heavy chain, ... | | Authors: | Cerutti, G, Verardi, R, Roederer, M, Shapiro, L. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DUN
 
 | | Cryo-EM Structure of Antibody SKW11 in complex with Western Equine Encephalitis Virus spike (local refinement from VLP particles) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody SKW11 heavy chain, ... | | Authors: | Cerutti, G, Verardi, R, Roederer, M, Shapiro, L. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (5.84 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
5ZJQ
 
 | | Structure of AbdB/Exd complex bound to a 'Red14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*TP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Baburajendran, N, Zeiske, T, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
8EM4
 
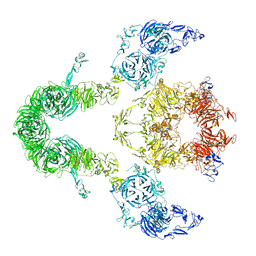 | | Cryo-EM structure of LRP2 at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Brasch, J, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
8EM7
 
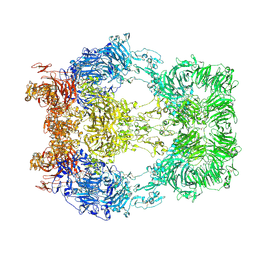 | | Cryo-EM structure of LRP2 at pH 5.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
5ZJR
 
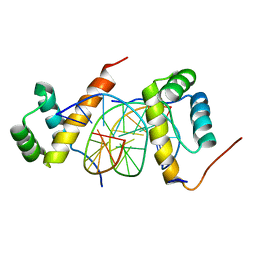 | | Structure of AbdB/Exd complex bound to a 'Magenta14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*TP*CP*GP*TP*AP*AP*AP*TP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*AP*TP*TP*TP*AP*CP*GP*AP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Zeiske, T, Baburajendran, N, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
5ZJT
 
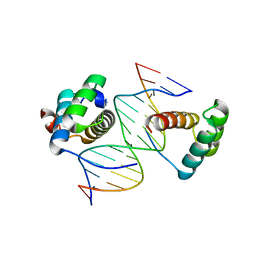 | | Structure of AbdB/Exd complex bound to a 'Black14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*AP*AP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*TP*TP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Zeiske, T, Baburajendran, N, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
5ZJS
 
 | | Structure of AbdB/Exd complex bound to a 'Blue14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*TP*AP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*TP*AP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Baburajendran, N, Zeiske, T, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
8TNU
 
 | | Cryo-EM structure of TRNM-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Roark, R.S, Morano, N.C, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TO7
 
 | | Cryo-EM structure of HERH-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HERH-b*01 heavy chain, ... | | Authors: | Roark, R.S, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TO9
 
 | | Cryo-EM structure of TRNM-f*01 Fab in complex with HIV-1 Env trimer ConC SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Roark, R.S, Morano, N.C, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
1Y7E
 
 | |
1YRH
 
 | |
1YDG
 
 | |
1Y65
 
 | |
