3CWO
 
 | | A beta/alpha-barrel built by the combination of fragments from different folds | | Descriptor: | SULFATE ION, beta/alpha-barrel protein based on 1THF and 1TMY | | Authors: | Bharat, T.A.M, Eisenbeis, S, Zeth, K, Hocker, B. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A beta alpha-barrel built by the combination of fragments from different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5NV6
 
 | | Structure of human transforming growth factor beta-induced protein (TGFBIp). | | Descriptor: | ACETATE ION, Transforming growth factor-beta-induced protein ig-h3 | | Authors: | Garcia-Castellanos, R, Nielsen, S.N, Runager, K, Thogersen, B.I, Goulas, T, Enghild, J.J, Gomis-Ruth, F.X. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural and Functional Implications of Human Transforming Growth Factor beta-Induced Protein, TGFBIp, in Corneal Dystrophies.
Structure, 25, 2017
|
|
3DJ5
 
 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 290. | | Descriptor: | 3-({3-[(6-amino-5-bromopyrimidin-4-yl)sulfanyl]propanoyl}amino)-4-methoxy-N-phenylbenzamide, serine/threonine kinase 6 | | Authors: | Elling, R.A, Erlanson, D.A, Yang, W, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
3DJ6
 
 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 823. | | Descriptor: | 4-methoxy-N-phenyl-3-({3-[(1H-pyrrolo[2,3-b]pyridin-5-ylmethyl)sulfanyl]propanoyl}amino)benzamide, serine/threonine kinase 6 | | Authors: | Elling, R.A, Erlanson, D.A, Yang, W, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
2XDH
 
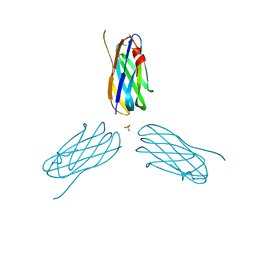 | | Non-cellulosomal cohesin from the hyperthermophilic archaeon Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, COHESIN, MAGNESIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Noach, I, Borovok, I, Kwiat, M, Rosenheck, S, Shimon, L.J.W, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Non-Cellulosomal Cohesin from the Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Proteins, 79, 2011
|
|
1TYJ
 
 | | Crystal Structure Analysis of type II Cohesin A11 from Bacteroides cellulosolvens | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, cellulosomal scaffoldin | | Authors: | Noach, I, Frolow, F, Jakoby, H, Rosenheck, S, Shimon, L.J.W, Lamed, R, Bayer, E.A. | | Deposit date: | 2004-07-08 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements
J.Mol.Biol., 348, 2005
|
|
2AYP
 
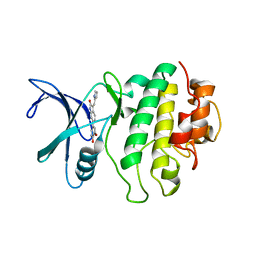 | | Crystal Structure of CHK1 with an Indol Inhibitor | | Descriptor: | (3Z)-6-(4-HYDROXY-3-METHOXYPHENYL)-3-(1H-PYRROL-2-YLMETHYLENE)-1,3-DIHYDRO-2H-INDOL-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Lin, N.-H, Xia, P, Kovar, P, Chen, Z, Zhang, H, Rosenberg, S.H, Sham, H.L. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and biological evaluation of 3-ethylidene-1,3-dihydro-indol-2-ones as novel checkpoint 1 inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2J4Q
 
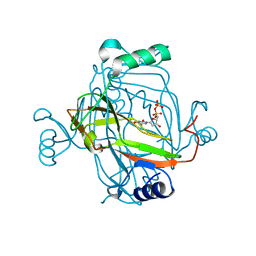 | | Crystal structure of a E138A Escherichia coli dCTP deaminase mutant enzyme in complex with dTTP | | Descriptor: | DEOXYCYTIDINE TRIPHOSPHATE DEAMINASE, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Johansson, E, Thymark, M, Bynck, J.H, Fanoe, M, Larsen, S, Willemoes, M. | | Deposit date: | 2006-09-05 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of Dctp Deaminase from Escherichia Coli by Nonallosteric Dttp Binding to an Inactive Form of the Enzyme
FEBS J., 274, 2007
|
|
1OGH
 
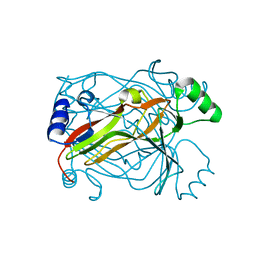 | |
2J4H
 
 | | Crystal structure of a H121A Escherichia coli dCTP deaminase mutant enzyme | | Descriptor: | DEOXYCYTIDINE DIPHOSPHATE, DEOXYCYTIDINE TRIPHOSPHATE DEAMINASE, MAGNESIUM ION | | Authors: | Johansson, E, Thymark, M, Bynck, J.H, Fanoe, M, Larsen, S, Willemoes, M. | | Deposit date: | 2006-08-31 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Regulation of Dctp Deaminase from Escherichia Coli by Nonallosteric Dttp Binding to an Inactive Form of the Enzyme
FEBS J., 274, 2007
|
|
1YI5
 
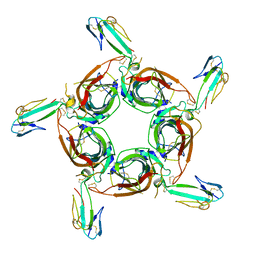 | | Crystal structure of the a-cobratoxin-AChBP complex | | Descriptor: | Acetylcholine-binding protein, Long neurotoxin 1 | | Authors: | Bourne, Y, Talley, T.T, Hansen, S.B, Taylor, P, Marchot, P. | | Deposit date: | 2005-01-11 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors
Embo J., 24, 2005
|
|
6GYV
 
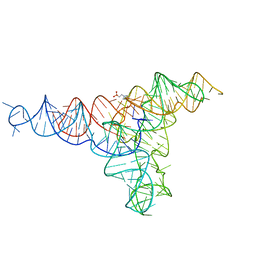 | | Lariat-capping ribozyme (circular permutation form) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lariat-capping ribozyme, MAGNESIUM ION, ... | | Authors: | Masquida, B, Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E. | | Deposit date: | 2018-07-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.50003624 Å) | | Cite: | Speciation of a group I intron into a lariat capping ribozyme.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
3DJ7
 
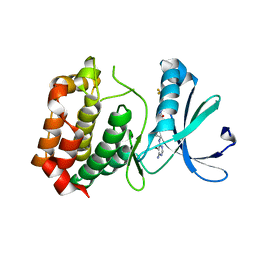 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 130. | | Descriptor: | 1-(5-{2-[(6-amino-5-bromopyrimidin-4-yl)amino]ethyl}-1,3-thiazol-2-yl)-3-[3-(trifluoromethyl)phenyl]urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Yang, W, Erlanson, D.A, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
1AK8
 
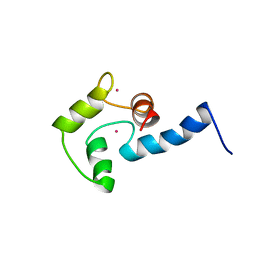 | | NMR SOLUTION STRUCTURE OF CERIUM-LOADED CALMODULIN AMINO-TERMINAL DOMAIN (CE2-TR1C), 23 STRUCTURES | | Descriptor: | CALMODULIN, CERIUM (III) ION | | Authors: | Bentrop, D, Bertini, I, Cremonini, M.A, Forsen, S, Luchinat, C, Malmendal, A. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the paramagnetic complex of the N-terminal domain of calmodulin with two Ce3+ ions by 1H NMR.
Biochemistry, 36, 1997
|
|
2VN0
 
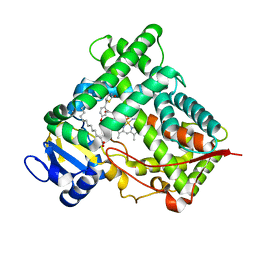 | | CYP2C8DH COMPLEXED WITH TROGLITAZONE | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, CYTOCHROME P450 2C8, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Sansen, S, Stout, C.D, Johnson, E.F. | | Deposit date: | 2008-01-30 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J. Biol. Chem., 283, 2008
|
|
1GGW
 
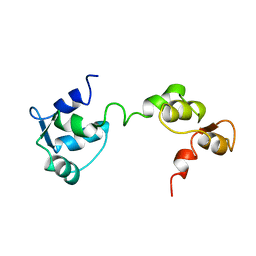 | |
1NI1
 
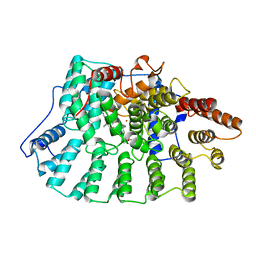 | | Imidazole and cyanophenyl farnesyl transferase inhibitors | | Descriptor: | 2-CHLORO-5-(3-CHLORO-PHENYL)-6-[(4-CYANO-PHENYL)-(3-METHYL-3H-IMIDAZOL-4-YL)- METHOXYMETHYL]-NICOTINONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Tong, Y, Lin, N.H, Wang, L, Hasvold, L, Wang, W, Leonard, N, Li, T, Li, Q, Cohen, J, Gu, W.Z, Zhang, H, Stoll, V, Bauch, J, Marsh, K, Rosenberg, S.H, Sham, H.L. | | Deposit date: | 2002-12-20 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of potent imidazole and cyanophenyl containing farnesyltransferase inhibitors with improved oral bioavailability.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1KCY
 
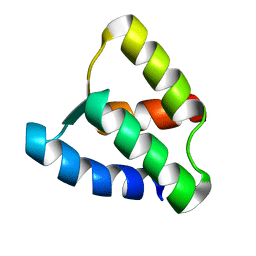 | | NMR solution structure of apo calbindin D9k (F36G + P43M mutant) | | Descriptor: | calbindin D9k | | Authors: | Nelson, M.R, Thulin, E, Fagan, P.A, Forsen, S, Chazin, W.J. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The EF-hand domain: a globally cooperative structural unit.
Protein Sci., 11, 2002
|
|
2ARF
 
 | | Solution structure of the Wilson ATPase N-domain in the presence of ATP | | Descriptor: | WILSON DISEASE ATPASE | | Authors: | Dmitriev, O, Tsivkovskii, R, Abildgaard, F, Morgan, C.T, Markley, J.L, Lutsenko, S. | | Deposit date: | 2005-08-19 | | Release date: | 2006-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-domain of Wilson disease protein: Distinct nucleotide-binding environment and effects of disease mutations
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1H4H
 
 | | Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Sabini, E, Wilson, K.S, Danielsen, S, Schulein, M, Davies, G.J. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis and Specificity in Enzymatic Glycoside Hydrolysis: A 2,5B Conformation for the Glycosyl-Enzyme Intermediate Revealed by the Structure of the Bacillus Agaradhaerens Family 11 Xylanase.
Chem.Biol., 6, 1999
|
|
1UWZ
 
 | | Bacillus subtilis cytidine deaminase with an Arg56 - Ala substitution | | Descriptor: | CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ZINC ION | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
1UX0
 
 | | Bacillus subtilis cytidine deaminase with an Arg56 - Gln substitution | | Descriptor: | CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ZINC ION | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
1UX1
 
 | | Bacillus subtilis cytidine deaminase with a Cys53His and an Arg56Gln substitution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ... | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
3KJ6
 
 | | Crystal structure of a Methylated beta2 Adrenergic Receptor-Fab complex | | Descriptor: | Beta-2 adrenergic receptor, Fab heavy chain, Fab light chain, ... | | Authors: | Bokoch, M.P, Zou, Y, Rasmussen, S.G.F, Liu, C.W, Nygaard, R, Rosenbaum, D.M, Fung, J.J, Choi, H.-J, Thian, F.S, Kobilka, T.S, Puglisi, J.D, Weis, W.I, Pardo, L, Prosser, S, Mueller, L, Kobilka, B.K. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor.
Nature, 463, 2010
|
|
1ETP
 
 | |
