3KQ4
 
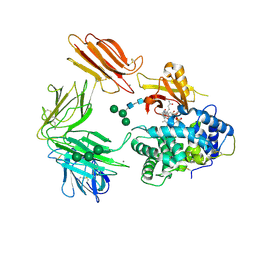 | | Structure of Intrinsic Factor-Cobalamin bound to its receptor Cubilin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Andersen, C.B.F, Madsen, M, Moestrup, S.K, Andersen, G.R. | | Deposit date: | 2009-11-17 | | Release date: | 2010-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes.
Nature, 464, 2010
|
|
7AAV
 
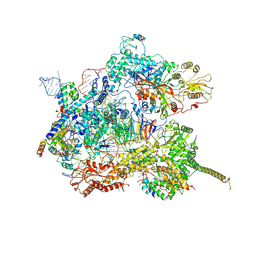 | | Human pre-Bact-2 spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, D-chiro inositol hexakisphosphate, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
4USR
 
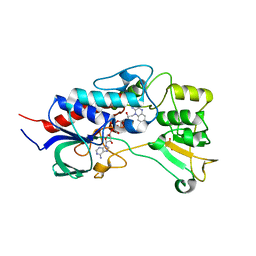 | | Structure of flavin-containing monooxygenase from Pseudomonas stutzeri NF13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MONOOXYGENASE | | Authors: | Jensen, C.N, Ali, S.T, Allen, M.J, Grogan, G. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Exploring Nicotinamide Cofactor Promiscuity in Nad(P)H-Dependent Flavin Containing Monooxygenases (Fmos) Using Natural Variation within the Phosphate Binding Loop. Structure and Activity of Fmos from Cellvibrio Sp. Br and Pseudomonas Stutzeri NF13
J.Mol.Catal., 109, 2014
|
|
6UJ6
 
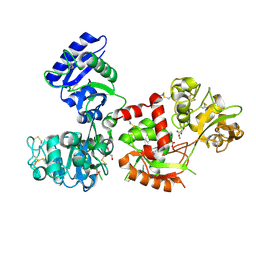 | | X-ray Crystal Structure of Chromium-transferrin with Synergistic Anion Malonate | | Descriptor: | BICARBONATE ION, CHROMIUM ION, GLYCEROL, ... | | Authors: | Petersen, C.M, Edwards, K.C, Gilbert, N.C, Vincent, J.B, Thompson, M.K. | | Deposit date: | 2019-10-02 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | X-ray structure of chromium(III)-containing transferrin: First structure of a physiological Cr(III)-binding protein.
J.Inorg.Biochem., 210, 2020
|
|
1M9Z
 
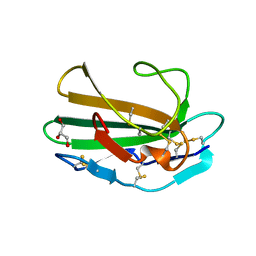 | | CRYSTAL STRUCTURE OF HUMAN TGF-BETA TYPE II RECEPTOR LIGAND BINDING DOMAIN | | Descriptor: | GLYCEROL, TGF-BETA RECEPTOR TYPE II | | Authors: | Boesen, C.C, Radaev, S, Motyka, S.A, Patamawenu, A, Sun, P.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-09-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | THE 1.1A CRYSTAL STRUCTURE OF HUMAN TGF-BETA TYPE II RECEPTOR LIGAND BINDING DOMAIN
Structure, 10, 2002
|
|
9EUO
 
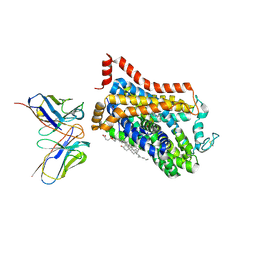 | | Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 2024
|
|
9EUP
 
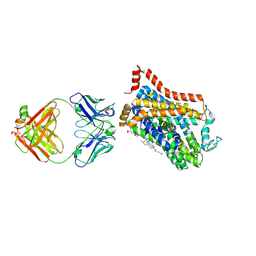 | | Inhibitor-free outward-open structure of Drosophila dopamine transporter | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 2024
|
|
4USQ
 
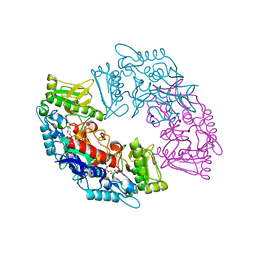 | | Structure of flavin-containing monooxygenase from Cellvibrio sp. BR | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PYRIDINE NUCLEOTIDE-DISULFIDE OXIDOREDUCTASE | | Authors: | Jensen, C.N, Ali, S.T, Allen, M.J, Grogan, G. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Exploring Nicotinamide Cofactor Promiscuity in Nad(P)H-Dependent Flavin Containing Monooxygenases (Fmos) Using Natural Variation within the Phosphate Binding Loop. Structure and Activity of Fmos from Cellvibrio Sp. Br and Pseudomonas Stutzeri NF13
J.Mol.Catal., 109, 2014
|
|
9G11
 
 | |
4F4O
 
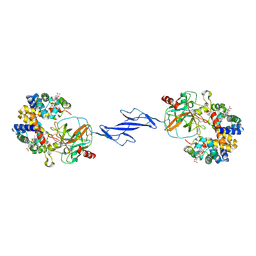 | | Structure of the Haptoglobin-Haemoglobin Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin, ... | | Authors: | Andersen, C.B.F, Torvund-Jensen, M, Nielsen, M.J, Oliveira, C.L.P, Hersleth, H.P, Andersen, N.H, Pedersen, J.S, Andersen, G.R, Moestrup, S.K. | | Deposit date: | 2012-05-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the haptoglobin-haemoglobin complex.
Nature, 489, 2012
|
|
6GJE
 
 | | Structure of the Amnionless(20-357)-Cubilin(36-135) complex | | Descriptor: | Cubilin, Protein amnionless | | Authors: | Larsen, C, Etzerodt, A, Madsen, M, Skjoedt, K, Moestrup, S.K, Andersen, C.B.F. | | Deposit date: | 2018-05-16 | | Release date: | 2018-12-19 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural assembly of the megadalton-sized receptor for intestinal vitamin B12uptake and kidney protein reabsorption.
Nat Commun, 9, 2018
|
|
7ABG
 
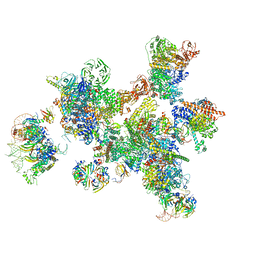 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ABF
 
 | | Human pre-Bact-1 spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ABI
 
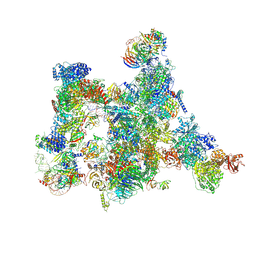 | | Human pre-Bact-2 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Beta-catenin-like protein 1, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ABH
 
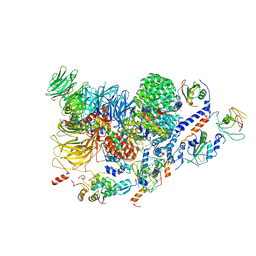 | | Human pre-Bact-2 spliceosome (SF3b/U2 snRNP portion) | | Descriptor: | Cell division cycle 5-like protein, DNA/RNA-binding protein KIN17, MINX M3 pre-mRNA, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
8P1I
 
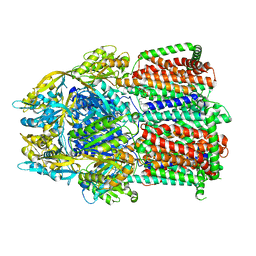 | | Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL), 3-chloranyl-2,6-di(piperazin-4-ium-1-yl)quinoline, Efflux pump membrane transporter, ... | | Authors: | Boernsen, C, Mueller, R.T, Pos, K.M, Frangakis, A.S. | | Deposit date: | 2023-05-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Pyridylpiperazine efflux pump inhibitor boosts in vivo antibiotic efficacy against K. pneumoniae.
Embo Mol Med, 16, 2024
|
|
4QF9
 
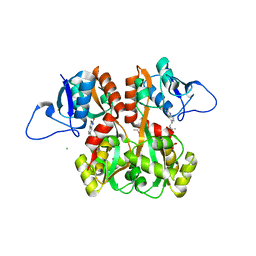 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid at 2.28 A resolution | | Descriptor: | (2S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kristensen, C.M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2014-05-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Binding Mode of an alpha-Amino Acid-Linked Quinoxaline-2,3-dione Analogue at Glutamate Receptor Subtype GluK1.
ACS Chem Neurosci, 6, 2015
|
|
3GWK
 
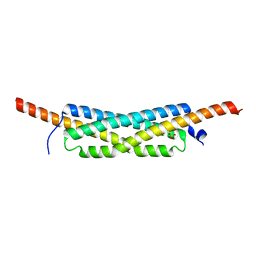 | | Structure of the homodimeric WXG-100 family protein from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein SAG1039, SULFATE ION | | Authors: | Poulsen, C, Gries, F, Wilmanns, M, Song, Y.H. | | Deposit date: | 2009-04-01 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern.
Plos One, 9, 2014
|
|
4FH1
 
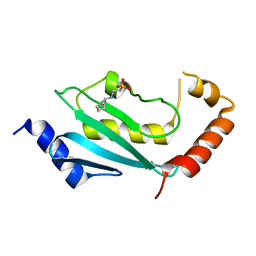 | | S. cerevisiae Ubc13-N79A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Ubiquitin-conjugating enzyme E2 13 | | Authors: | Berndsen, C.E, Wolberger, C, Wiener, R. | | Deposit date: | 2012-06-05 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A conserved asparagine has a structural role in ubiquitin-conjugating enzymes.
Nat.Chem.Biol., 9, 2013
|
|
3GVM
 
 | |
7ZS6
 
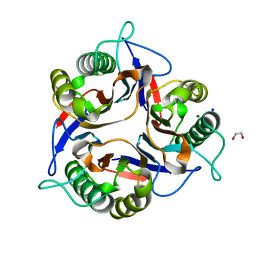 | | Crystal structure of Apis mellifera RidA | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Reactive intermediate deaminase A, ... | | Authors: | Visentin, C, Rizzi, G, Ricagno, S. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Apis mellifera RidA, a novel member of the canonical YigF/YER057c/UK114 imine deiminase superfamily of enzymes pre-empting metabolic damage.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
4KTD
 
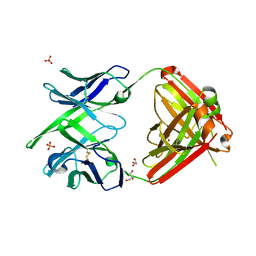 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE136, from non-human primate | | Descriptor: | GE136 Heavy Chain Fab, GE136 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Standfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KTE
 
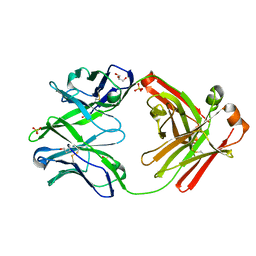 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate | | Descriptor: | GE148 Heavy Chain Fab, GE148 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Stanfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J9P
 
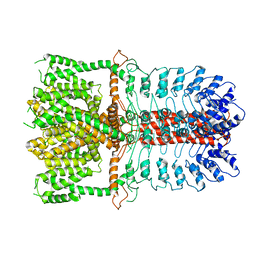 | | Structure of the TRPA1 ion channel determined by electron cryo-microscopy | | Descriptor: | Maltose-binding periplasmic protein, Transient receptor potential cation channel subfamily A member 1 chimera | | Authors: | Paulsen, C.E, Armache, J.-P, Gao, Y, Cheng, Y, Julius, D. | | Deposit date: | 2015-02-14 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structure of the TRPA1 ion channel suggests regulatory mechanisms.
Nature, 520, 2015
|
|
3GWM
 
 | |
