1XBT
 
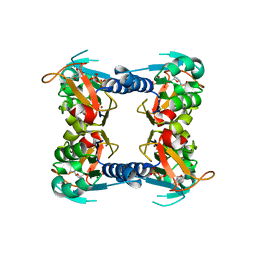 | | Crystal Structure of Human Thymidine Kinase 1 | | Descriptor: | MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE, Thymidine kinase, ... | | Authors: | Welin, M, Kosinska, U, Mikkelsen, N.E, Carnrot, C, Zhu, C, Wang, L, Eriksson, S, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2004-08-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of thymidine kinase 1 of human and mycoplasmic origin
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2C7U
 
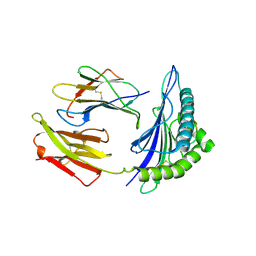 | | Conflicting selective forces affect CD8 T-cell receptor contact sites in an HLA-A2 immunodominant HIV epitope. | | Descriptor: | BETA-2-MICROGLOBULIN, GAG PROTEIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Iversen, A.K, Stewart-Jones, G, Learn, G.H, Christie, N, Sylvester-Hviid, C, Armitage, A.E, Kaul, R, Beattie, T, Lee, J.K, Li, Y, Chotiyarnwong, P, Dong, T, Xu, X, Luscher, M.A, MacDonald, K, Ullum, H, Klarlund-Pedersen, B, Skinhoj, P, Fugger, J.L, Buus, S, Mullins, J.I, Jones, E.Y, van der Merwe, P.A, McMichael, A.J. | | Deposit date: | 2005-11-29 | | Release date: | 2006-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Conflicting Selective Forces Affect T Cell Receptor Contacts in an Immunodominant Human Immunodeficiency Virus Epitope.
Nat.Immunol., 7, 2006
|
|
1XJ9
 
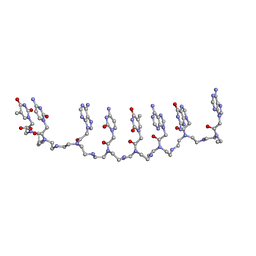 | | Crystal structure of a partly self-complementary peptide nucleic acid (PNA) oligomer showing a duplex-triplex network | | Descriptor: | peptide nucleic acid, (H-P(*GPN*TPN*APN*GPN*APN*TPN*CPN*APN*CPN*TPN)-LYS-NH2) | | Authors: | Petersson, B, Nielsen, B.B, Rasmussen, H, Larsen, I.K, Gajhede, M, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2004-09-23 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Partly Self-Complementary Peptide Nucleic Acid (PNA) Oligomer Showing a Duplex-Triplex Network
J.Am.Chem.Soc., 127, 2005
|
|
2WVJ
 
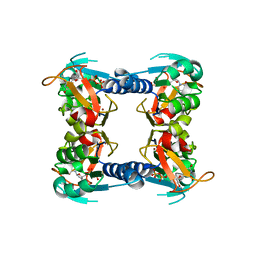 | | Mutation of Thr163 to Ser in Human Thymidine Kinase Shifts the Specificity from Thymidine towards the Nucleoside Analogue Azidothymidine | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, CYTOSOLIC, ... | | Authors: | Skovgaard, T, Uhlin, U, Munch-Petersen, B. | | Deposit date: | 2009-10-17 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative Active-Site Mutation Study of Human and Caenorhabditis Elegans Thymidine Kinase 1.
FEBS J., 279, 2012
|
|
5DFG
 
 | |
2BNU
 
 | | Structural and kinetic basis for heightened immunogenicity of T cell vaccines | | Descriptor: | T-CELL RECEPTOR ALPHA CHAIN C REGION, T-CELL RECEPTOR BETA CHAIN C REGION | | Authors: | Chen, J.-L, Stewart-Jones, G, Bossi, G, Lissin, N.M, Wooldridge, L, Choi, E.M.L, Held, G, Dunbar, P.R, Esnouf, R.M, Sami, M, Boultier, J.M, Rizkallah, P.J, Renner, C, Sewell, A, Van Der Merwe, P.A, Jackobsen, B.K, Griffiths, G, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2005-04-04 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Kinetic Basis for Heightened Immunogenicity of T Cell Vaccines.
J.Exp.Med., 201, 2005
|
|
4FR0
 
 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with SAM | | Descriptor: | Arsenic methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Ajees, A.A, Marapakala, K, Packianathan, C, Sankaran, B, Rosen, B.P. | | Deposit date: | 2012-06-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Biochemistry, 51, 2012
|
|
5DRH
 
 | | Crystal structure of apo C-As lyase | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase, SODIUM ION | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of apo C-As lyase
To Be Published
|
|
1S5O
 
 | | Structural and Mutational Characterization of L-carnitine Binding to Human carnitine Acetyltransferase | | Descriptor: | CARNITINE, carnitine acetyltransferase isoform 2 | | Authors: | Govindasamy, L, Kukar, T, Lian, W, Pedersen, B, Gu, Y, Agbandje-Mckenna, M, Jin, S, Mckenna, R, Wu, D. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutational characterization of l-carnitine binding to human carnitine acetyltransferase.
J.Struct.Biol., 146, 2004
|
|
9ETN
 
 | | Crystal structure of murine CRTAC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Cartilage acidic protein 1, ... | | Authors: | Beugelink, J.W, Hof, H, Janssen, B.J.C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | CRTAC1 has a Compact beta-propeller-TTR Core Stabilized by Potassium Ions.
J.Mol.Biol., 436, 2024
|
|
4FSD
 
 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with As(III) | | Descriptor: | ARSENIC, Arsenic methyltransferase, CALCIUM ION, ... | | Authors: | Ajees, A.A, Marapakala, K, Packianathan, C, Sankaran, B, Rosen, B.P. | | Deposit date: | 2012-06-27 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Biochemistry, 51, 2012
|
|
1FU6
 
 | |
4YKF
 
 | | Crystal Structure of the Alkylhydroperoxide Reductase subunit F (AhpF) with NADH from Escherichia coli | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alkyl hydroperoxide reductase subunit F, CADMIUM ION, ... | | Authors: | Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps
Biochim.Biophys.Acta, 1847, 2015
|
|
4YKG
 
 | | Crystal Structure of the Alkylhydroperoxide Reductase subunit F (AhpF) with NAD+ from Escherichia coli | | Descriptor: | Alkyl hydroperoxide reductase subunit F, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps
Biochim.Biophys.Acta, 1847, 2015
|
|
4QL7
 
 | | Crystal structure of C-terminus truncated Alkylhydroperoxide Reductase subunit C (AhpC1-172) from E. coli | | Descriptor: | Alkylhydroperoxide Reductase subunit C | | Authors: | Kamariah, N, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2014-06-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Key roles of the Escherichia coli AhpC C-terminus in assembly and catalysis of alkylhydroperoxide reductase, an enzyme essential for the alleviation of oxidative stress.
Biochim.Biophys.Acta, 1837, 2014
|
|
4BYG
 
 | | ATPase crystal structure | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Pedersen, B.P, Morth, J.P, Wang, J, Gourdon, P, Nissen, P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dephosphorylation of Pib-Type Cu(I)-Atpases as Studied by Metallofluoride Complexes
To be Published
|
|
6QPP
 
 | | Rhizomucor miehei lipase propeptide complex, native | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
6QPR
 
 | | Rhizomucor miehei lipase propeptide complex, Ser95/Ile96 deletion mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
3TGL
 
 | | STRUCTURE AND MOLECULAR MODEL REFINEMENT OF RHIZOMUCOR MIEHEI TRIACYLGLYCERIDE LIPASE: A CASE STUDY OF THE USE OF SIMULATED ANNEALING IN PARTIAL MODEL REFINEMENT | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURE AND MOLECULAR-MODEL REFINEMENT OF RHIZOMUCOR-MIEHEI TRIACYLGLYCERIDE LIPASE - A CASE-STUDY OF THE USE OF SIMULATED ANNEALING IN PARTIAL MODEL REFINEMENT.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1SYI
 
 | | X-RAY STRUCTURE OF THE Y702F MUTANT OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 2.1 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1SYH
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
2JJ8
 
 | |
2JCS
 
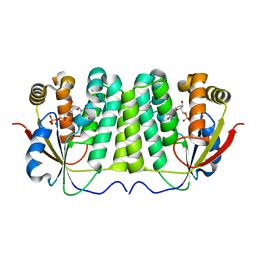 | | Active site mutant of dNK from D. melanogaster with dTTP bound | | Descriptor: | DEOXYNUCLEOSIDE KINASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Egeblad-Welin, L, Sonntag, Y, Eklund, H, Munch-Petersen, B. | | Deposit date: | 2007-01-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Studies of Active-Site Mutants from Drosophila Melanogaster Deoxyribonucleoside Kinase. Investigations of the Putative Catalytic Glutamate- Arginine Pair and of Residues Responsible for Substrate Specificity.
FEBS J., 274, 2007
|
|
7ZJ5
 
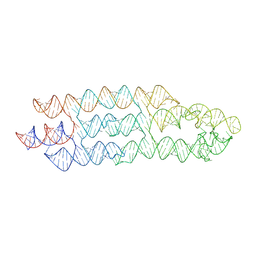 | | Unbound state of a brocolli-pepper aptamer FRET tile. | | Descriptor: | POTASSIUM ION, brocolli-pepper aptamer | | Authors: | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
6RK8
 
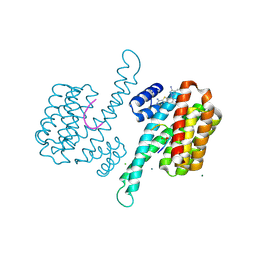 | | Fragment AZ-014 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-(3-azanyl-4-methyl-pyrazol-1-yl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
