4XSV
 
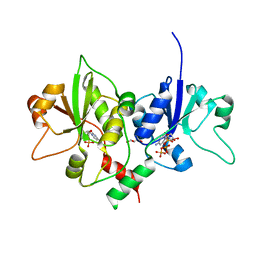 | |
3G0H
 
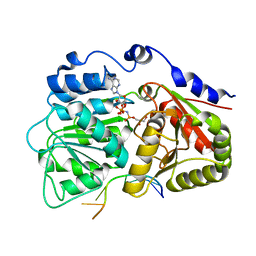 | | Human dead-box RNA helicase DDX19, in complex with an ATP-analogue and RNA | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase DDX19B, GLYCEROL, ... | | Authors: | Karlberg, T, Lehtio, L, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The DEXD/H-box RNA Helicase DDX19 Is Regulated by an {alpha}-Helical Switch.
J.Biol.Chem., 284, 2009
|
|
4UXB
 
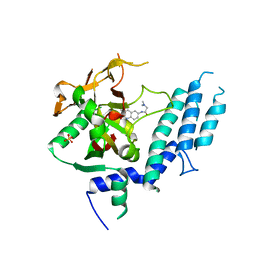 | | Human ARTD1 (PARP1) - Catalytic domain in complex with inhibitor PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, POLY ADP-RIBOSE POLYMERASE 1, SULFATE ION | | Authors: | Tresaugues, L, Thorsell, A.G, Karlberg, T, Schuler, H. | | Deposit date: | 2014-08-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4R5W
 
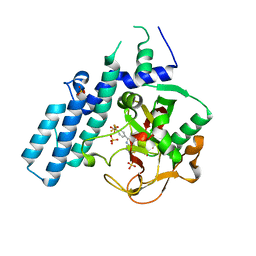 | | Human artd1 (parp1) - catalytic domain in complex with inhibitor xav939 | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Karlberg, T, Thorsell, A.G, Schuler, H. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J.Med.Chem., 60, 2017
|
|
4R6E
 
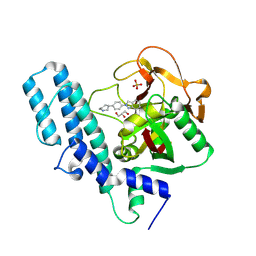 | | Human artd1 (parp1) - catalytic domain in complex with inhibitor niraparib | | Descriptor: | 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Karlberg, T, Thorsell, A.G, Brock, J, Schuler, H. | | Deposit date: | 2014-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J.Med.Chem., 60, 2017
|
|
4X52
 
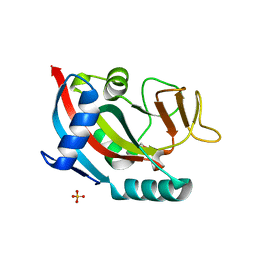 | | Human PARP13 (ZC3HAV1), C-Terminal PARP Domain (H810N; N830Y variant) | | Descriptor: | GLYCEROL, SULFATE ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Karlberg, T, Thorsell, A.G, Klepsch, M, Schuler, H. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
6FZM
 
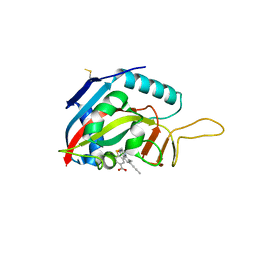 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor ITK6 | | Descriptor: | 4-[(8-methyl-4-oxidanylidene-7-prop-1-ynyl-3~{H}-quinazolin-2-yl)methylsulfanyl]benzoic acid, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Kirby, I.T, Sreenivasan, R, Cohen, M.S, Schuler, H. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity.
Cell Chem Biol, 25, 2018
|
|
6G0W
 
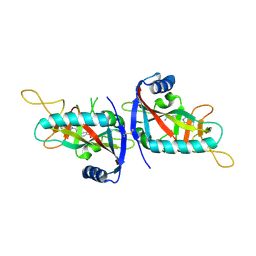 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor MCD72 | | Descriptor: | 4-[3-[4-(4-fluorophenyl)piperidin-1-yl]carbonylphenoxy]benzamide, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Holechek, J, Lease, R, Ferraris, D, Schuler, H. | | Deposit date: | 2018-03-20 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design, synthesis and evaluation of potent and selective inhibitors of mono-(ADP-ribosyl)transferases PARP10 and PARP14.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6FX7
 
 | | Crystal structure of in vitro evolved Af1521 | | Descriptor: | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, [Protein ADP-ribosylglutamate] hydrolase AF_1521 | | Authors: | Karlberg, T, Thorsell, A.G, Nowak, K, Hottiger, M.O, Schuler, H. | | Deposit date: | 2018-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Engineering Af1521 improves ADP-ribose binding and identification of ADP-ribosylated proteins.
Nat Commun, 11, 2020
|
|
6FXI
 
 | | Human PARP10 (ARTD10), catalytic fragment in complex with 3-aminobenzamide and citrate | | Descriptor: | 3-aminobenzamide, CITRIC ACID, GLYCEROL, ... | | Authors: | Karlberg, T, Thorsell, A.G, Schuler, H. | | Deposit date: | 2018-03-09 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human PARP10 (ARTD10), catalytic fragment in complex with 3-aminobenzamide and citrate
To Be Published
|
|
6GNK
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form bound to Carba-NAD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 14-3-3 protein beta/alpha, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GNJ
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form in complex with STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
4RV6
 
 | |
4TVJ
 
 | | HUMAN ARTD2 (PARP2) - CATALYTIC DOMAIN IN COMPLEX WITH OLAPARIB | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Pinto, A.F, Schuler, H. | | Deposit date: | 2014-06-27 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4UND
 
 | | HUMAN ARTD1 (PARP1) - CATALYTIC DOMAIN IN COMPLEX WITH INHIBITOR TALAZOPARIB | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, POLY [ADP-RIBOSE] POLYMERASE 1, SODIUM ION | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Klepsch, M, Pinto, A.F, Tresaugues, L, Moche, M, Schuler, H. | | Deposit date: | 2014-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
2J4E
 
 | | THE ITP COMPLEX OF HUMAN INOSINE TRIPHOSPHATASE | | Descriptor: | INOSINE 5'-TRIPHOSPHATE, INOSINE TRIPHOSPHATE PYROPHOSPHATASE, INOSINIC ACID, ... | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmbergschiavone, L, Hogbom, M, Kotenyova, T, Landry, R, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Schuler, H, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-08-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Inosine Triphosphatase: Substrate Binding and Implication of the Inosine Triphosphatase Deficiency Mutation P32T.
J.Biol.Chem., 282, 2007
|
|
4MXE
 
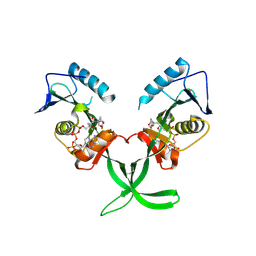 | | Human ESCO1 (Eco1/Ctf7 ortholog), acetyltransferase domain in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase ESCO1 | | Authors: | Karlberg, T, Wisniewska, M, Thorsell, A.G, Kouznetsova, E, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Svensson, L, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-26 | | Release date: | 2015-04-08 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sister Chromatid Cohesion Establishment Factor ESCO1 Operates by Substrate-Assisted Catalysis.
Structure, 24, 2016
|
|
5QHU
 
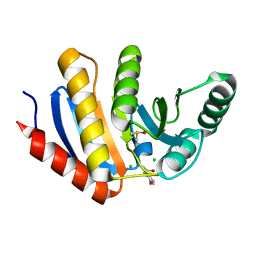 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMSOA000341b | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N-(2-hydroxyphenyl)acetamide, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI4
 
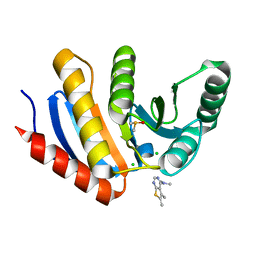 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000466a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QHW
 
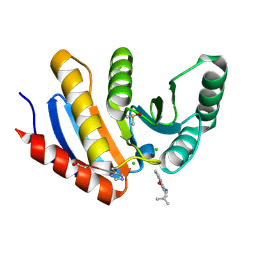 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000347a | | Descriptor: | 2-methyl-~{N}-(2-methylpropyl)imidazo[1,2-a]pyridine-3-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI0
 
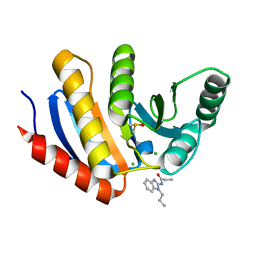 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000352a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI3
 
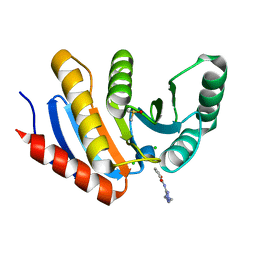 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000475a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI5
 
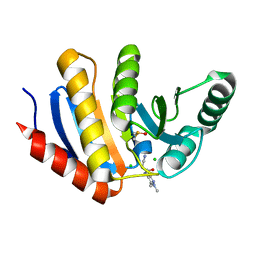 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000633a | | Descriptor: | 2-cyano-~{N}-(1,3,5-trimethylpyrazol-4-yl)ethanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI6
 
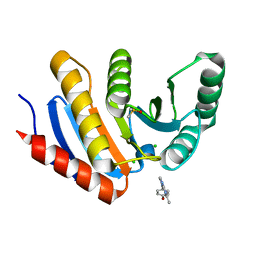 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000597a | | Descriptor: | 4-[(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)amino]phenol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QI1
 
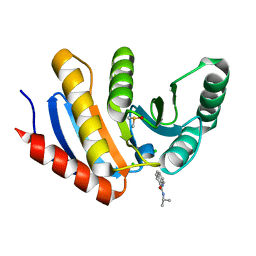 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000474a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
