4E6U
 
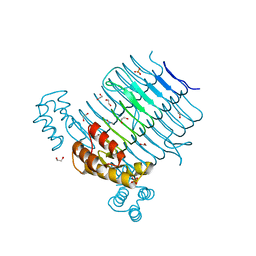 | | Structure of LpxA from Acinetobacter baumannii at 1.4A resolution (P63 form) | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7RMY
 
 | | De Novo designed tunable protein pockets, D_3-337 | | Descriptor: | De Novo designed tunable homodimer, D_3-337 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RMX
 
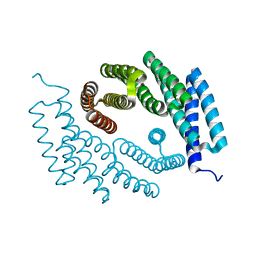 | | Structure of De Novo designed tunable symmetric protein pockets | | Descriptor: | Tunable symmetric protein, D_3_212 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7K2X
 
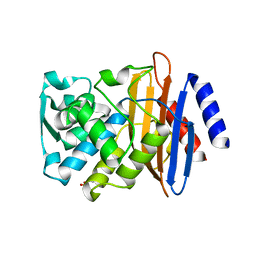 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase | | Descriptor: | Beta-lactamase, GLYCEROL | | Authors: | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | Deposit date: | 2020-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
4FSD
 
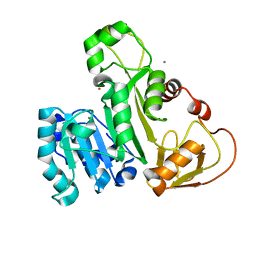 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with As(III) | | Descriptor: | ARSENIC, Arsenic methyltransferase, CALCIUM ION, ... | | Authors: | Ajees, A.A, Marapakala, K, Packianathan, C, Sankaran, B, Rosen, B.P. | | Deposit date: | 2012-06-27 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Biochemistry, 51, 2012
|
|
4E75
 
 | | Structure of LpxD from Acinetobacter baumannii at 2.85A resolution (P21 form) | | Descriptor: | UDP-3-O-acylglucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure determination of LpxD from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4FR0
 
 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with SAM | | Descriptor: | Arsenic methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Ajees, A.A, Marapakala, K, Packianathan, C, Sankaran, B, Rosen, B.P. | | Deposit date: | 2012-06-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Biochemistry, 51, 2012
|
|
7LLH
 
 | | KPC-2 F72Y mutant with acylated imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
4GV5
 
 | | X-ray structure of crotamine, a cell-penetrating peptide from the Brazilian snake Crotalus durissus terrificus | | Descriptor: | Crotamine Ile-19, GLYCEROL, SULFATE ION, ... | | Authors: | Coronado, M.A, Gabdulkhakov, A, Georgieva, D, Sankaran, B, Murakami, M.T, Arni, R.K, Betzel, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the polypeptide crotamine from the Brazilian rattlesnake Crotalus durissus terrificus.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4E6T
 
 | | Structure of LpxA from Acinetobacter baumannii at 1.8A resolution (P212121 form) | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CITRATE ANION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7S7M
 
 | | Complex of tissue inhibitor of metalloproteinases-1 (TIMP-1) mutant (L34G/M66D/T98G/P131S/Q153N) with matrix metalloproteinase-3 catalytic domain (MMP-3cd) | | Descriptor: | CALCIUM ION, Metalloproteinase inhibitor 1, Stromelysin-1, ... | | Authors: | Coban, M, Raeeszadeh-Sarmazdeh, M, Sankaran, B, Hockla, A, Radisky, E.S. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering of tissue inhibitor of metalloproteinases TIMP-1 for fine discrimination between closely related stromelysins MMP-3 and MMP-10.
J.Biol.Chem., 298, 2022
|
|
4KU8
 
 | | Structures of PKGI Reveal a cGMP-Selective Activation Mechanism | | Descriptor: | GLYCINE, cGMP-dependent Protein Kinase 1 | | Authors: | Huang, G.Y, Kim, J.J, Reger, A.S, Lorenz, R, Moon, E.W, Casteel, D.E, Sankaran, B, Herberg, F.W, Kim, C. | | Deposit date: | 2013-05-21 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Structural Basis for Cyclic-Nucleotide Selectivity and cGMP-Selective Activation of PKG I.
Structure, 22, 2014
|
|
7L99
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1302 | | Descriptor: | Bromodomain testis-specific protein, N-[3-(acetylamino)-4-methylphenyl]-3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carboxamide, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Sharma, R, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L9A
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1102 | | Descriptor: | BETA-MERCAPTOETHANOL, Bromodomain testis-specific protein, N~1~-(5-{[3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carbonyl]amino}-2-methylphenyl)-N~4~-methylbenzene-1,4-dicarboxamide | | Authors: | Sharma, R, Kaur, G, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4IN2
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | C-like protease | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4INH
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | DIMETHYL SULFOXIDE, Genome polyprotein, peptide inhibitor, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
7S7L
 
 | | Complex of tissue inhibitor of metalloproteinases-1 (TIMP-1) mutant (L34G/M66S/E67Y/L133N/S155L) with matrix metalloproteinase-3 catalytic domain (MMP-3cd) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Coban, M, Raeeszadeh-Sarmazdeh, M, Hockla, A, Sankaran, B, Radisky, E.S. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Engineering of tissue inhibitor of metalloproteinases TIMP-1 for fine discrimination between closely related stromelysins MMP-3 and MMP-10.
J.Biol.Chem., 298, 2022
|
|
7S5S
 
 | | CTX-M-15 WT in complex with BLIP WT | | Descriptor: | Beta-lactamase, Beta-lactamase inhibitory protein | | Authors: | Lu, S, Palzkill, T, Hu, L.Y, Prasad, B.V.V, Sankaran, B. | | Deposit date: | 2021-09-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An active site loop toggles between conformations to control antibiotic hydrolysis and inhibition potency for CTX-M beta-lactamase drug-resistance enzymes.
Nat Commun, 13, 2022
|
|
3PMO
 
 | | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2010-11-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3QHY
 
 | | Structural, thermodynamic and kinetic analysis of the picomolar binding affinity interaction of the beta-lactamase inhibitor protein-II (BLIP-II) with class A beta-lactamases | | Descriptor: | Beta-lactamase, Beta-lactamase inhibitory protein II | | Authors: | Brown, N.G, Chow, D.C, Sankaran, B, Zwart, P, Prasad, B.V.V, Palzkill, T, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2011-01-26 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
J.Biol.Chem., 286, 2011
|
|
3SJP
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid, ZINC ION | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
6WMK
 
 | | Crystal structure of beta sheet heterodimer LHD29 | | Descriptor: | Beta sheet heterodimer LHD29 - Chain A, Beta sheet heterodimer LHD29 - Chain B | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2020-04-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
7RSW
 
 | | Crystal structure of group B human rotavirus VP8* | | Descriptor: | Outer capsid protein VP4, peptide | | Authors: | Hu, L, Salmen, W, Sankaran, B, Prasad, B.V. | | Deposit date: | 2021-08-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography.
Commun Biol, 5, 2022
|
|
3SLD
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to A trisaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SI9
 
 | | Crystal structure of Dihydrodipicolinate Synthase from Bartonella Henselae | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate synthase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Staker, B.L, Abendroth, J, Sankaran, B. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cloning, expression, purification, crystallization and X-ray diffraction analysis of dihydrodipicolinate synthase from the human pathogenic bacterium Bartonella henselae strain Houston-1 at 2.1 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
