3GWJ
 
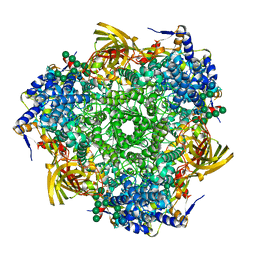 | | Crystal structure of Antheraea pernyi arylphorin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylphorin, FORMIC ACID, ... | | Authors: | Ryu, K.S, Lee, J.O, Kwon, T.H, Kim, S. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin
Biochem.J., 421, 2009
|
|
3KYF
 
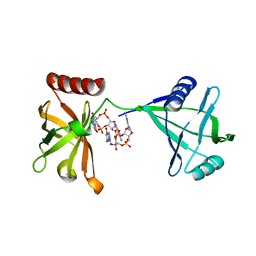 | | Crystal structure of P4397 complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
5YA2
 
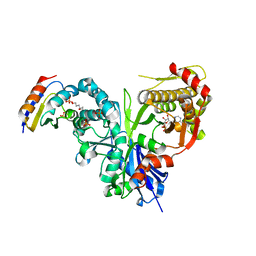 | | Crystal structure of LsrK-HPr complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Autoinducer-2 kinase, HEXANE-1,6-DIOL, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-30 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
1X8D
 
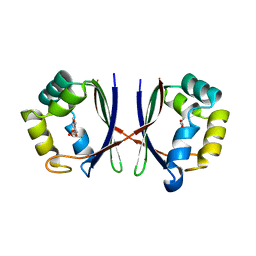 | | Crystal structure of E. coli YiiL protein containing L-rhamnose | | Descriptor: | Hypothetical protein yiiL, L-RHAMNOSE | | Authors: | Ryu, K.S, Kim, J.I, Cho, S.J, Park, D, Park, C, Lee, J.O, Choi, B.S. | | Deposit date: | 2004-08-18 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Monosaccharide Specificity of Escherichia coli Rhamnose Mutarotase
J.Mol.Biol., 349, 2005
|
|
3KYG
 
 | | Crystal structure of VCA0042 (L135R) complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein VCA0042 | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
6L5K
 
 | | ARF5 Aux/IAA17 Complex | | Descriptor: | Auxin response factor 5, Auxin-responsive protein IAA17 | | Authors: | Ryu, K.S, Suh, J.Y, Cha, S.Y, Kim, Y.I, Park, C.K. | | Deposit date: | 2019-10-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Determinants of PB1 Domain Interactions in Auxin Response Factor ARF5 and Repressor IAA17.
J.Mol.Biol., 432, 2020
|
|
1P1A
 
 | | NMR structure of ubiquitin-like domain of hHR23B | | Descriptor: | UV excision repair protein RAD23 homolog B | | Authors: | Ryu, K.S, Lee, K.J, Bae, S.H, Kim, B.K, Kim, K.A, Choi, B.S. | | Deposit date: | 2003-04-11 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a
J.Biol.Chem., 278, 2003
|
|
5YA1
 
 | | crystal structure of LsrK-HPr complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autoinducer-2 kinase, HEXANE-1,6-DIOL, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
5YA0
 
 | | Crystal structure of LsrK and HPr complex | | Descriptor: | Autoinducer-2 kinase, HEXANE-1,6-DIOL, PHOSPHATE ION, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
4L4Y
 
 | |
4L5I
 
 | |
4L51
 
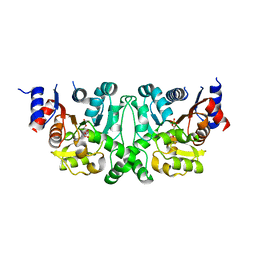 | |
4L5J
 
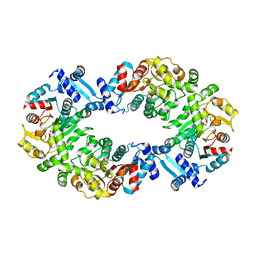 | |
4L4Z
 
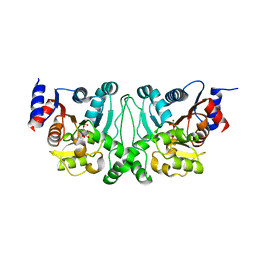 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | Descriptor: | (2S)-2,3,3-trihydroxy-4-oxopentyl dihydrogen phosphate, Transcriptional regulator LsrR | | Authors: | Ryu, K.S, Ha, J.H, Eo, Y. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
4L50
 
 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | Descriptor: | (2S)-2,3,3-trihydroxy-6-methyl-4-oxoheptyl dihydrogen phosphate, Transcriptional regulator LsrR | | Authors: | Ryu, K.S, Ha, J.H, Eo, Y. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
7X89
 
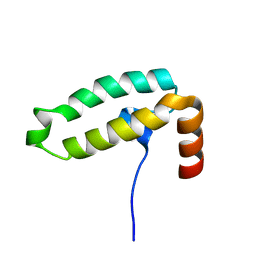 | | Tid1 | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Jang, J, Lee, S.H, Kang, D.H, Sim, D.W, Jo, K.S, Ryu, H, Kim, E.H, Ryu, K.S, Lee, J.H, Kim, J.H, Won, H.S. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the J-domain and GF-motif of the mitochondrial Hsp40, Tid1
To Be Published
|
|
2LJP
 
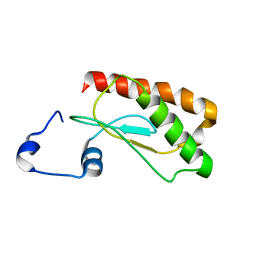 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for E.coli Ribonuclease P protein | | Descriptor: | Ribonuclease P protein component | | Authors: | Shin, J, Kim, K, Ryu, K, Han, K, Lee, Y, Choi, B. | | Deposit date: | 2011-09-21 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Escherichia coli C5 protein
To be Published
|
|
8K4K
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Olsalazine carbon derivative | | Descriptor: | 5-[(E)-2-(3-carboxy-4-oxidanyl-phenyl)ethenyl]-2-oxidanyl-benzoic acid, Flavin reductase (NADPH), GLYCEROL, ... | | Authors: | Ha, J.H, Jung, H.M, dela Cerna, M.V.C, Burlison, J.A, Lee, D, Ryu, K.S. | | Deposit date: | 2023-07-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Innovative Inhibitor with a New Chemical Moiety Aimed at Biliverdin IX beta Reductase for Thrombocytopenia and Resilient against Cellular Degradation.
Pharmaceutics, 16, 2024
|
|
2LSI
 
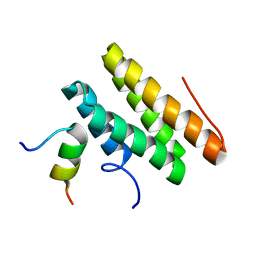 | |
6ILU
 
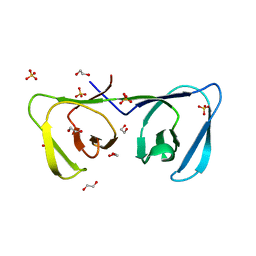 | | Endolysin LysPBC5 CBD | | Descriptor: | 1,2-ETHANEDIOL, Lysin, SULFATE ION | | Authors: | Suh, J.Y, Ryu, K.S, Ryu, S, Lee, K.O, Kong, M.S, Bae, J.W, Kim, I.T. | | Deposit date: | 2018-10-19 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural Basis for Cell-Wall Recognition by Bacteriophage PBC5 Endolysin.
Structure, 27, 2019
|
|
6M2D
 
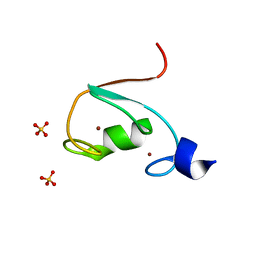 | | MUL1-RING domain | | Descriptor: | Mitochondrial ubiquitin ligase activator of NFKB 1, SULFATE ION, ZINC ION | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
6M2C
 
 | |
7BOL
 
 | | ubiquitin-conjugating enzyme, Ube2D2 | | Descriptor: | Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
4EXT
 
 | |
2KCC
 
 | | Solution Structure of biotinoyl domain from human acetyl-CoA carboxylase 2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Lee, C, Cheong, H, Ryu, K, Lee, J, Lee, W, Jeon, Y, Cheong, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
Proteins, 72, 2008
|
|
