3MG5
 
 | | Core-streptavidin mutant F130L in complex with biotin | | Descriptor: | BIOTIN, GLYCEROL, Streptavidin | | Authors: | Le Trong, I, Baugh, L, Stayton, P.S, Lybrand, T.P, Stenkamp, R.E. | | Deposit date: | 2010-04-05 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A distal point mutation in the streptavidin-biotin complex preserves structure but diminishes binding affinity: experimental evidence of electronic polarization effects?
Biochemistry, 49, 2010
|
|
4ZCE
 
 | | Crystal Structure of the dust mite allergen Der p 23 from Dermatophagoides pteronyssinus | | Descriptor: | 1,2-ETHANEDIOL, Dust mite allergen | | Authors: | Pedersen, L.C, Mueller, G.A, Randall, T.A, Glesner, J, Perera, L, Edwards, L.L, Chapman, M.D, London, R.E, Pomes, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-11-25 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Serological, genomic and structural analyses of the major mite allergen Der p 23.
Clin Exp Allergy, 46, 2016
|
|
5L9E
 
 | | CRYSTAL STRUCTURE OF HUMAN CARBONIC ANHYDRASE II IN COMPLEX WITH A QUINOLINE OLIGOAMIDE FOLDAMER | | Descriptor: | 8-azanyl-4-(2-hydroxy-2-oxoethyloxy)quinoline-2-carboxylic acid, 8-azanyl-4-(2-methylpropoxy)quinoline-2-carboxylic acid, Carbonic anhydrase 2, ... | | Authors: | Vallade, M, Langlois d'Estaintot, B, Granier, T, Huc, I. | | Deposit date: | 2016-06-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CRYSTAL STRUCTURE OF HUMAN CARBONIC ANHYDRASE II IN COMPLEX WITH A QUINOLINE OLIGOAMIDE FOLDAMER
To Be Published
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
1WS3
 
 | | Urate oxidase from aspergillus flavus complexed with uracil | | Descriptor: | URACIL, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Vivares, D, Bonnete, F, Castro, B, El Hajji, M, Prange, T. | | Deposit date: | 2004-10-29 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5L3O
 
 | | Crystal Structure of Human Carbonic Anhydrase II in Complex with a Quinoline Oligoamide Foldamer | | Descriptor: | 8-azanyl-4-(2-hydroxy-2-oxoethyloxy)quinoline-2-carboxylic acid, 8-azanyl-4-(2-methylpropoxy)quinoline-2-carboxylic acid, 8-azanyl-4-(3-azanylpropoxy)quinoline-2-carboxylic acid, ... | | Authors: | Jewginski, M, Langlois d'Estaintot, B, Granier, T, Huc, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
1OJJ
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
5LVS
 
 | | Self-assembled protein-aromatic foldamer complexes with 2:3 and 2:2:1 stoichiometries | | Descriptor: | 8-azanyl-4-(2-hydroxy-2-oxoethyloxy)quinoline-2-carboxylic acid, 8-azanyl-4-(2-methylpropoxy)quinoline-2-carboxylic acid, 8-azanyl-4-(3-azanylpropoxy)quinoline-2-carboxylic acid, ... | | Authors: | Jewginski, M, LANGLOIS D'ESTAINTOT, B, Granier, T, Huc, Y. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
1P3R
 
 | | Crystal structure of the phosphotyrosin binding domain(PTB) of mouse Disabled 1(Dab1) | | Descriptor: | Disabled homolog 2 | | Authors: | Yun, M, Keshvara, L, Park, C.G, Zhang, Y.M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.W. | | Deposit date: | 2003-04-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2.
J.Biol.Chem., 278, 2003
|
|
6P0Z
 
 | | Crystal structure of N-acetylated KRAS (2-169) bound to GDP and Mg | | Descriptor: | ACETYL GROUP, DI(HYDROXYETHYL)ETHER, GTPase KRas, ... | | Authors: | Dharmaiah, S, Tran, T.H, Yan, W, Simanshu, D.K. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.011 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
6PNY
 
 | | X-ray Structure of Flpp3 | | Descriptor: | Flpp3 | | Authors: | Zook, J.D, Shekhar, M, Hansen, D.T, Conrad, C, Grant, T.D, Gupta, C, White, T, Barty, A, Basu, S, Zhao, Y, Zatsepin, N.A, Ishchenko, A, Batyuk, A, Gati, C, Li, C, Galli, L, Coe, J, Hunter, M, Liang, M, Weierstall, U, Nelson, G, James, D, Stauch, B, Craciunescu, F, Thifault, D, Liu, W, Cherezov, V, Singharoy, A, Fromme, P. | | Deposit date: | 2019-07-03 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | XFEL and NMR Structures of Francisella Lipoprotein Reveal Conformational Space of Drug Target against Tularemia.
Structure, 28, 2020
|
|
1OS0
 
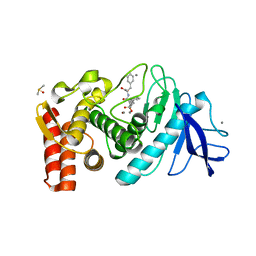 | | Thermolysin with an alpha-amino phosphinic inhibitor | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N-{(2R)-3-[(S)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Selkti, M, Tomas, A, Prange, T. | | Deposit date: | 2003-03-18 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions of a new alpha-aminophosphinic derivative inside the active site of TLN (thermolysin): a model for zinc-metalloendopeptidase inhibition.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1FE0
 
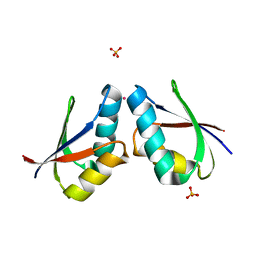 | | CRYSTAL STRUCTURE OF CADMIUM-HAH1 | | Descriptor: | CADMIUM ION, COPPER TRANSPORT PROTEIN ATOX1, SULFATE ION, ... | | Authors: | Wernimont, A.K, Huffman, D.L, Lamb, A.L, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2000-07-20 | | Release date: | 2001-01-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins.
Nat.Struct.Biol., 7, 2000
|
|
1OQN
 
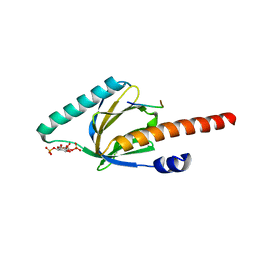 | | Crystal structure of the phosphotyrosine binding domain (PTB) of mouse Disabled 1 (Dab1) | | Descriptor: | Alzheimer's disease amyloid A4 protein homolog, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1 | | Authors: | Yun, M, Keshvara, L, Park, C.-G, Zhang, Y.-M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.-W. | | Deposit date: | 2003-03-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2
J.Biol.Chem., 278, 2003
|
|
3OBP
 
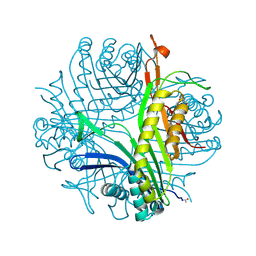 | | Anaerobic complex of urate oxidase with uric acid | | Descriptor: | SODIUM ION, URIC ACID, Uricase | | Authors: | Gabison, L, Chopard, C, Colloc'h, N, El Hajji, M, Castro, B, Chiadmi, M, Prange, T. | | Deposit date: | 2010-08-08 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray, ESR, and quantum mechanics studies unravel a spin well in the cofactor-less urate oxidase.
Proteins, 79, 2011
|
|
1WS2
 
 | | urate oxidase from aspergillus flavus complexed with 5,6-diaminouracil | | Descriptor: | 5,6-DIAMINOPYRIMIDINE-2,4(1H,3H)-DIONE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Vivares, D, Bonnete, F, Castro, B, El Hajji, M, Prange, T. | | Deposit date: | 2004-10-29 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7OGC
 
 | | GTPase HRAS under 650 MPa pressure | | Descriptor: | GTPase HRas, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Colloc'h, N.C, Girard, E, Prange, T, Kalbitzer, H.R. | | Deposit date: | 2021-05-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Equilibria between conformational states of the Ras oncogene protein revealed by high pressure crystallography.
Chem Sci, 13, 2022
|
|
7OGA
 
 | | GTPase HRAS under 200 MPa pressure | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Colloc'h, N.C, Kalbitzer, H.R, Girard, E, Prange, T. | | Deposit date: | 2021-05-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Equilibria between conformational states of the Ras oncogene protein revealed by high pressure crystallography.
Chem Sci, 13, 2022
|
|
7OGD
 
 | | GTPase HRAS mutant D33K under ambient pressure | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Colloc'h, N.C, Girard, E, Prange, T, Kalbitzer, H.R. | | Deposit date: | 2021-05-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Equilibria between conformational states of the Ras oncogene protein revealed by high pressure crystallography.
Chem Sci, 13, 2022
|
|
7OG9
 
 | | GTPase HRAS under ambient pressure | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Colloc'h, N, Girard, E, Prange, T, Kalbitzer, H.R. | | Deposit date: | 2021-05-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Equilibria between conformational states of the Ras oncogene protein revealed by high pressure crystallography.
Chem Sci, 13, 2022
|
|
7OGB
 
 | | GTPase HRAS under 500 MPa pressure | | Descriptor: | GTPase HRas, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Colloc'h, N.C, Girard, E, Prange, T, Kalbitzer, H.R. | | Deposit date: | 2021-05-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Equilibria between conformational states of the Ras oncogene protein revealed by high pressure crystallography.
Chem Sci, 13, 2022
|
|
7OGF
 
 | | GTPase HRAS mutant D33K under 900 MPa pressure | | Descriptor: | GTPase HRas, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Colloc'h, N.C, Girard, E, Prange, T, Kalbitzer, H.R. | | Deposit date: | 2021-05-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Equilibria between conformational states of the Ras oncogene protein revealed by high pressure crystallography.
Chem Sci, 13, 2022
|
|
7OGE
 
 | | GTPase HRAS mutant D33K under 200 MPa pressure | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Colloc'h, N.C, Girard, E, Prange, T, Kalbitzer, H.R. | | Deposit date: | 2021-05-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Equilibria between conformational states of the Ras oncogene protein revealed by high pressure crystallography.
Chem Sci, 13, 2022
|
|
5O1K
 
 | |
5JUP
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure II (mid-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
