1FBB
 
 | |
1FBK
 
 | |
2F9U
 
 | | HCV NS3 protease domain with NS4a peptide and a ketoamide inhibitor with a P2 norborane | | 分子名称: | 1,1-DIMETHYLETHYL [1-CYCLOHEXYL-2-[3-[[[1-[2-[[2-[[2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO]-2-OXOETHYL]AMINO]-1,2-DIOXOETHYL]PENTYL]AMINO]CARBONYL]-2-AZABICYCLO[2.2.1]HEPTAN-2-YL]-2-OXOETHYL]CARBAMATE, NS3 protease/helicase', ZINC ION, ... | | 著者 | Venkatraman, S, Njoroge, F.G, Wu, W, Girijavallabhan, V, Prongay, A.J, Butkiewicz, N, Pichardo, J. | | 登録日 | 2005-12-06 | | 公開日 | 2006-06-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Novel Inhibitors of Hepatitis C NS3-NS4A Serine Protease Derived from 2-Aza-bicyclo[2.2.1]heptane-3-carboxylic acid.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3KNX
 
 | | HCV NS3 protease domain with P1-P3 macrocyclic ketoamide inhibitor | | 分子名称: | (2R)-2-{(3S,13S,16aS,17aR,17bS)-13-[({(1S)-1-[(4,4-dimethyl-2,6-dioxopiperidin-1-yl)methyl]-2,2-dimethylpropyl}carbamoyl)amino]-17,17-dimethyl-1,14-dioxooctadecahydro-2H-cyclopropa[3,4]pyrrolo[1,2-a][1,4]diazacyclohexadecin-3-yl}-2-hydroxy-N-prop-2-en-1-ylethanamide, BETA-MERCAPTOETHANOL, HCV NS3 Protease, ... | | 著者 | Venkatraman, S, Velazquez, F, Wu, W, Blackman, M, Chen, K.X, Bogen, S, Nair, L, Tong, X, Chase, R, Hart, A, Agrawal, S, Pichardo, J, Prongay, A, Cheng, K.-C, Girijavallabhan, V, Piwinski, J, Shih, N.-Y, Njoroge, F.G. | | 登録日 | 2009-11-12 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Discovery and structure-activity relationship of P1-P3 ketoamide derived macrocyclic inhibitors of hepatitis C virus NS3 protease.
J.Med.Chem., 52, 2009
|
|
3CJI
 
 | |
3EYD
 
 | | Structure of HCV NS3-4A Protease with an Inhibitor Derived from a Boronic Acid | | 分子名称: | HCV NS3, HCV NS4a peptide, ZINC ION, ... | | 著者 | Venkatraman, S, Wu, W, Prongay, A.J, Girijavallabhan, V, Njoroge, F.G. | | 登録日 | 2008-10-20 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Potent inhibitors of HCV-NS3 protease derived from boronic acids.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7KD8
 
 | | TtgR C137I I141W M167L F168Y mutant in complex with resveratrol | | 分子名称: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | 著者 | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | 登録日 | 2020-10-08 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1A
 
 | | TtgR quadruple mutant (C137I I141W M167L F168Y) | | 分子名称: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION | | 著者 | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | 登録日 | 2020-09-07 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1C
 
 | | TtgR in complex with resveratrol | | 分子名称: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | 著者 | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | 登録日 | 2020-09-07 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
8ILU
 
 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | 分子名称: | (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-[2-(2-methyl-1,3-benzothiazol-6-yl)-1,2,4-triazol-3-yl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, SODIUM ION, ... | | 著者 | Kumar, A, Jinal, S, Raman, S, Ghosh, K. | | 登録日 | 2023-03-04 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Identification of Benzothiazole Derived Monosaccharides as Potent, Selective, and Orally Bioavailable Inhibitors of Human and Mouse Galectin-3
To Be Published
|
|
2L15
 
 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | 分子名称: | Cold shock protein CspA | | 著者 | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | 登録日 | 2010-07-22 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
7XFA
 
 | | Structure of human Galectin-3 CRD in complex with monosaccharide inhibitor | | 分子名称: | (2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-[2-[5-chloranyl-2-(trifluoromethyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | 著者 | Shukla, J, Raman, S, Ghosh, K. | | 登録日 | 2022-04-01 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Identification of Monosaccharide Derivatives as Potent, Selective, and Orally Bioavailable Inhibitors of Human and Mouse Galectin-3.
J.Med.Chem., 65, 2022
|
|
4UQK
 
 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | 著者 | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | 登録日 | 2014-06-24 | | 公開日 | 2014-08-13 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (16.4 Å) | | 主引用文献 | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
6R1T
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | 分子名称: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.02 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R25
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, H2B, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 実験手法 | ELECTRON MICROSCOPY (4.61 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
2XE0
 
 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | 分子名称: | 24MER DNA, ACETATE ION, I-CREI V2V3 VARIANT, ... | | 著者 | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Redondo, P, Villate, M, Merino, N, Marenchino, M, D'Abramo, M, Gervasio, F.L, Grizot, S, Daboussi, F, Smith, J, Chion-Sotine, I, Paques, F, Duchateau, P, Alibes, A, Stricher, F, Serrano, L, Blanco, F.J, Montoya, G. | | 登録日 | 2010-05-10 | | 公開日 | 2010-09-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Molecular Basis of Engineered Meganuclease Targeting of the Endogenous Human Rag1 Locus
Nucleic Acids Res., 39, 2011
|
|
5KUF
 
 | | GluK2EM with 2S,4R-4-methylglutamate | | 分子名称: | 2S,4R-4-METHYLGLUTAMATE, Glutamate receptor ionotropic, kainate 2 | | 著者 | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | 登録日 | 2016-07-13 | | 公開日 | 2016-09-07 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
5KUH
 
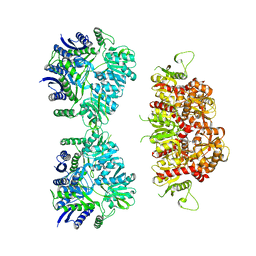 | | GluK2EM with LY466195 | | 分子名称: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | 著者 | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | 登録日 | 2016-07-13 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (11.6 Å) | | 主引用文献 | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
8SEU
 
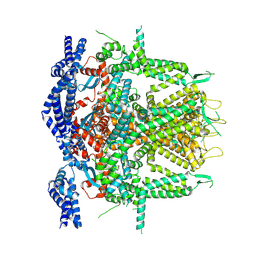 | | Cryo-EM Structure of RyR1 (Local Refinement of TMD) | | 分子名称: | Ryanodine receptor 1, ZINC ION | | 著者 | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | 登録日 | 2023-04-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SER
 
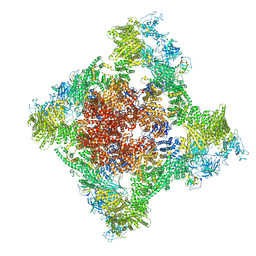 | | Cryo-EM Structure of RyR1 + Adenosine | | 分子名称: | ADENOSINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | 著者 | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | 登録日 | 2023-04-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEP
 
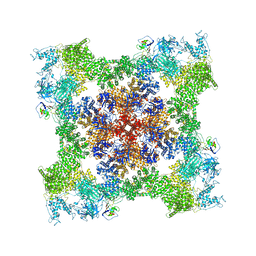 | | Cryo-EM Structure of RyR1 + ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | 著者 | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | 登録日 | 2023-04-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEV
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S (Local Refinement of TMD) | | 分子名称: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ZINC ION | | 著者 | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | 登録日 | 2023-04-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEQ
 
 | | Cryo-EM Structure of RyR1 + AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | 著者 | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | 登録日 | 2023-04-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEY
 
 | | Cryo-EM Structure of RyR1 + Adenosine (Local Refinement of TMD) | | 分子名称: | ADENOSINE, Ryanodine receptor 1, ZINC ION | | 著者 | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | 登録日 | 2023-04-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
