4LJ8
 
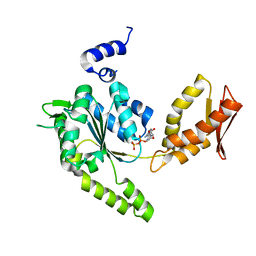 | | ClpB NBD2 R621Q from T. thermophilus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3C02
 
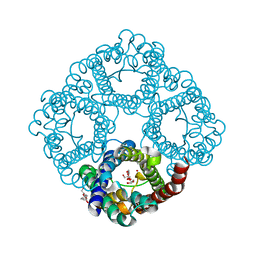 | |
2AE9
 
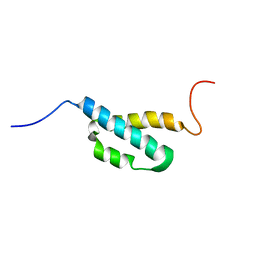 | | Solution Structure of the theta subunit of DNA polymerase III from E. coli | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Mueller, G.A, Kirby, T.W, Derose, E.F, Li, D, Schaaper, R.M, London, R.E. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of the Escherichia coli DNA Polymerase III {theta} Subunit.
J.Bacteriol., 187, 2005
|
|
6S8F
 
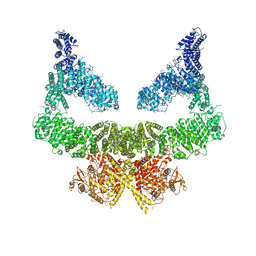 | | Structure of nucleotide-bound Tel1/ATM | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1 | | Authors: | Yates, L.A, Williams, R.M, Ayala, R, Zhang, X. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM Structure of Nucleotide-Bound Tel1ATMUnravels the Molecular Basis of Inhibition and Structural Rationale for Disease-Associated Mutations.
Structure, 28, 2020
|
|
4V5E
 
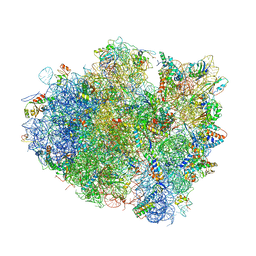 | | Insights into translational termination from the structure of RF2 bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Jin, H, Neubauer, C, Voorhees, R.M, Petry, S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-04-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Insights Into Translational Termination from the Structure of Rf2 Bound to the Ribosome.
Science, 322, 2008
|
|
6CCF
 
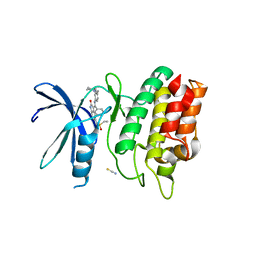 | | Crystal Structure of the Human CAMKK1A in complex with Hesperadin | | Descriptor: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase kinase 1, N-[2-OXO-3-((E)-PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]IMINO}METHYL)-2,6-DIHYDRO-1H-INDOL-5-YL]ETHANESULFONAMIDE, ... | | Authors: | Santiago, A.S, Counago, R.M, dos Reis, C.V, Ramos, P.Z, Silva, P.N.B, Drewry, D, Elkins, J.M, Massirer, K.B, Arruda, P, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human CAMKK1A in complex with Hesperadin
To be Published
|
|
1ZW9
 
 | |
5K01
 
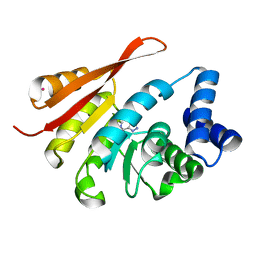 | | Crystal Structure of COMT in complex with 2,7-dimethyl-3-(1H-pyrazol-3-yl)imidazo[1,2-a]pyridine | | Descriptor: | 2,7-dimethyl-3-(1H-pyrazol-5-yl)imidazo[1,2-a]pyridine, Catechol O-methyltransferase, POTASSIUM ION | | Authors: | Ehler, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Crystal Structure of COMT
To Be Published
|
|
4LJ4
 
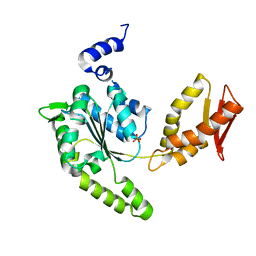 | | ClpB NBD2 from T. thermophilus, nucleotide-free | | Descriptor: | Chaperone protein ClpB, PHOSPHATE ION | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3NTI
 
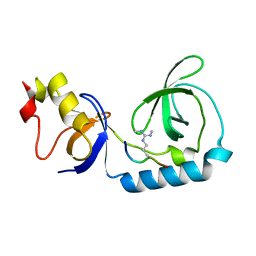 | | Crystal structure of Tudor and Aubergine [R15(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
4W4T
 
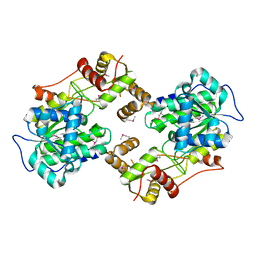 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | Descriptor: | ACETATE ION, MxaA | | Authors: | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
3NTK
 
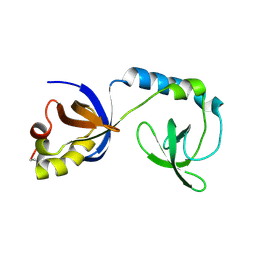 | | Crystal structure of Tudor | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
4V5D
 
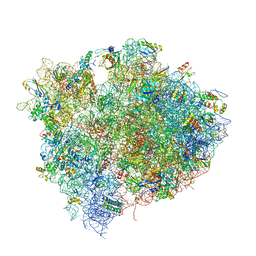 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
1N7U
 
 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM I | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
2AAZ
 
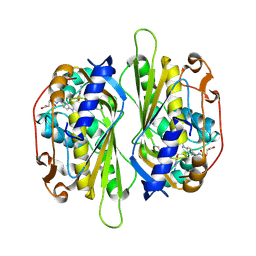 | | Cryptococcus neoformans thymidylate synthase complexed with substrate and an antifolate | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Finer-Moore, J.S, Anderson, A.C, O'Neil, R.H, Costi, M.P, Ferrari, S, Krucinski, J, Stroud, R.M. | | Deposit date: | 2005-07-14 | | Release date: | 2005-12-06 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of Cryptococcus neoformans thymidylate synthase suggests strategies for using target dynamics for species-specific inhibition.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4UXV
 
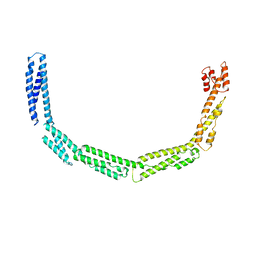 | | Cytoplasmic domain of bacterial cell division protein EzrA | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
4LJ7
 
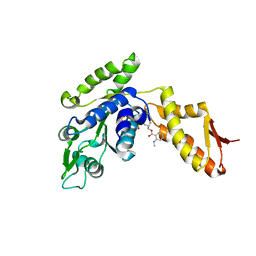 | | ClpB NBD2 K601Q from T. thermophilus in complex with MANT-dADP | | Descriptor: | 2'(3')-O-N-METHYLANTHRANILOYL-ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB, PHOSPHATE ION | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2KFX
 
 | |
4LK3
 
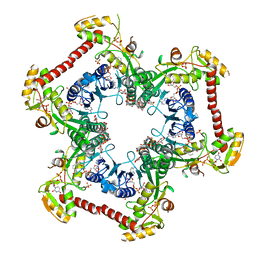 | | Crystal structure of Human UDP-xylose synthase R236A substitution | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYROPHOSPHATE 2-, SULFATE ION, ... | | Authors: | Walsh Jr, R.M, Polizzi, S.J, Wood, Z.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Man o' war mutation in UDP-alpha-D-xylose synthase favors the abortive catalytic cycle and uncovers a latent potential for hexamer formation.
Biochemistry, 54, 2015
|
|
8GF7
 
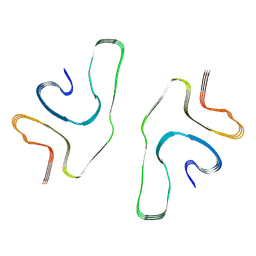 | | Cryo-EM structure of serine 87 O-GlcNAc-modified alpha-synuclein fibrils | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-synuclein | | Authors: | Balana, J.A, Nguyen, A.B, Saelices, L, Pratt, R.M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | O-GlcNAc forces an alpha-synuclein amyloid strain with notably diminished seeding and pathology.
Nat.Chem.Biol., 20, 2024
|
|
1NJE
 
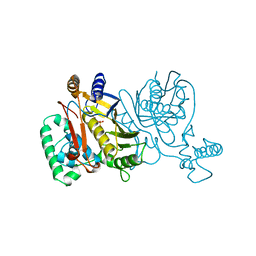 | |
2KWX
 
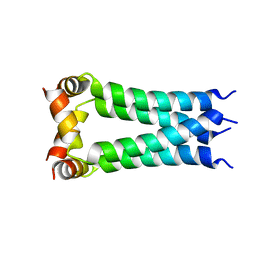 | |
1CER
 
 | |
4LJ6
 
 | | ClpB NBD2 from T. thermophilus in complex with AMPPCP | | Descriptor: | Chaperone protein ClpB, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LJ9
 
 | | ClpB NBD2 R621Q from T. thermophilus in complex with AMPPCP | | Descriptor: | Chaperone protein ClpB, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
