5H1M
 
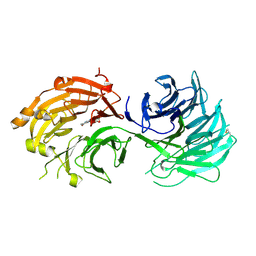 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with M7G | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Gem-associated protein 5 | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
7TN9
 
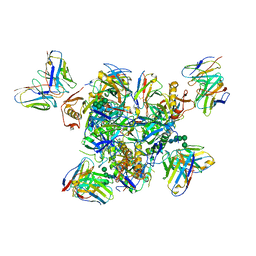 | | Structure of the Inmazeb cocktail and resistance to escape against Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, GP2, ... | | Authors: | Rayaprolu, V, Fulton, B, Rafique, A, Arturo, E, Williams, D, Hariharan, C, Callaway, H, Parvate, A, Schendel, S.L, Parekh, D, Hui, S, Shaffer, K, Pascal, K.E, Wloga, E, Giordano, S, Copin, R, Franklin, M, Boytz, R.M, Donahue, C, Davey, R, Baum, A, Kyratsous, C.A, Saphire, E.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the Inmazeb cocktail and resistance to Ebola virus escape.
Cell Host Microbe, 31, 2023
|
|
6XXG
 
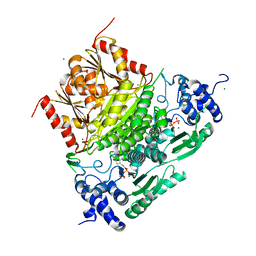 | | Structure of truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Deinococcus radiodurans | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-01-27 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties.
Biochem.Biophys.Res.Commun., 539, 2021
|
|
1DJ0
 
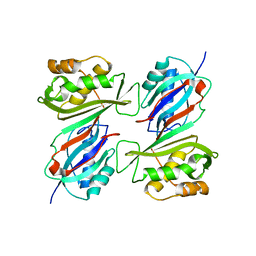 | | THE CRYSTAL STRUCTURE OF E. COLI PSEUDOURIDINE SYNTHASE I AT 1.5 ANGSTROM RESOLUTION | | Descriptor: | CHLORIDE ION, PSEUDOURIDINE SYNTHASE I | | Authors: | Foster, P.G, Huang, L, Santi, D.V, Stroud, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I.
Nat.Struct.Biol., 7, 2000
|
|
1DIY
 
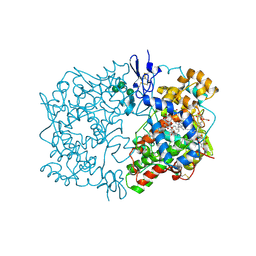 | | CRYSTAL STRUCTURE OF ARACHIDONIC ACID BOUND IN THE CYCLOOXYGENASE ACTIVE SITE OF PGHS-1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, ... | | Authors: | Malkowski, M.G, Ginell, S.L, Smith, W.L, Garavito, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The productive conformation of arachidonic acid bound to prostaglandin synthase.
Science, 289, 2000
|
|
6SF2
 
 | | Ternary complex of human bone morphogenetic protein 9 (BMP9) growth factor domain, its prodomain and extracellular domain of activin receptor-like kinase 1 (ALK1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Serine/threonine-protein kinase receptor R3 | | Authors: | Salmon, R.M, Guo, J, Yu, M, Li, W. | | Deposit date: | 2019-07-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis of ALK1-mediated signalling by BMP9/BMP10 and their prodomain-bound forms.
Nat Commun, 11, 2020
|
|
6SDV
 
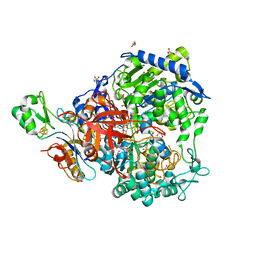 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Formate reduced form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
6SAL
 
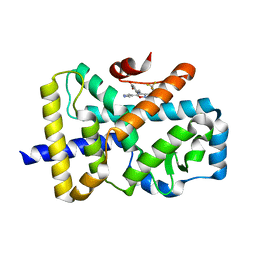 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM26 | | Descriptor: | 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Doveston, R.G, Brunsveld, L. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Ligand-Based Design of Allosteric Retinoic Acid Receptor-Related Orphan Receptor gamma t (ROR gamma t) Inverse Agonists.
J.Med.Chem., 63, 2020
|
|
7TXT
 
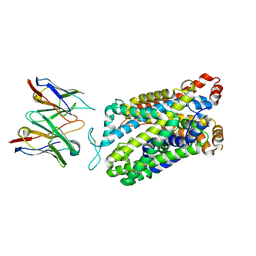 | | Structure of human serotonin transporter bound to small molecule '8090 in lipid nanodisc and NaCl | | Descriptor: | 1-[4-(4-fluorophenyl)-1,3-thiazol-2-yl]piperazine, 15B8 Fab heavy chain, 15B8 Fab light chain, ... | | Authors: | Singh, I, Seth, A, Billesboelle, C.B, Braz, J, Rodriguiz, R.M, Roy, K, Bekele, B, Craik, V, Huang, X.P, Boytsov, D, Lak, P, O'Donnell, H, Sandtner, W, Roth, B.L, Basbaum, A.I, Wetsel, W.C, Manglik, A, Shoichet, B.K, Rudnick, G. | | Deposit date: | 2022-02-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based discovery of conformationally selective inhibitors of the serotonin transporter.
Cell, 186, 2023
|
|
7U4A
 
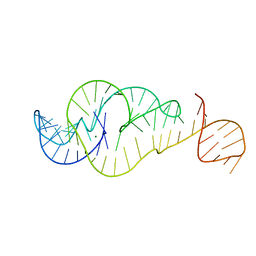 | | Crystal Structure of Zika virus xrRNA1 mutant | | Descriptor: | MAGNESIUM ION, RNA (70-MER) | | Authors: | Thompson, R.D, Carbaugh, D.L, Nielsen, J.R, Witt, C, Meganck, R.M, Rangadurai, A, Zhao, B, Bonin, J.P, Nathan, N.T, Marzluff, W.F, Frank, A.T, Lazear, H.M, Zhang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Dynamic Basis of Xrn1 Resistance in Mosquito-borne Flavivirus RNA
To Be Published
|
|
6SF1
 
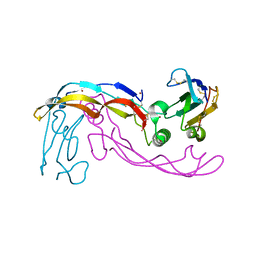 | | Bone morphogenetic protein 10 (BMP10) complexed with extracellular domain of activin receptor-like kinase 1 (ALK1). | | Descriptor: | Bone morphogenetic protein 10, NICKEL (II) ION, Serine/threonine-protein kinase receptor R3 | | Authors: | Salmon, R.M, Guo, J, Yu, M, Li, W. | | Deposit date: | 2019-07-30 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of ALK1-mediated signalling by BMP9/BMP10 and their prodomain-bound forms.
Nat Commun, 11, 2020
|
|
5EQG
 
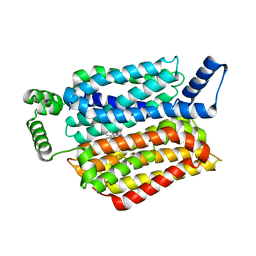 | | Human GLUT1 in complex with inhibitor (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide | | Descriptor: | (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1DJ9
 
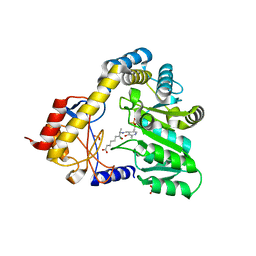 | | CRYSTAL STRUCTURE OF 8-AMINO-7-OXONANOATE SYNTHASE (OR 7-KETO-8AMINIPELARGONATE OR KAPA SYNTHASE) COMPLEXED WITH PLP AND THE PRODUCT 8(S)-AMINO-7-OXONANONOATE (OR KAPA). THE ENZYME OF BIOTIN BIOSYNTHETIC PATHWAY. | | Descriptor: | 8-AMINO-7-OXONONANOATE SYNTHASE, MAGNESIUM ION, N-[7-KETO-8-AMINOPELARGONIC ACID]-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
6XA2
 
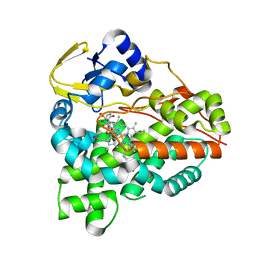 | | Structure of the tirandamycin C-bound P450 monooxygenase TamI | | Descriptor: | (3E)-3-{(2E,4E,6R)-1-hydroxy-4-methyl-6-[(1R,3R,4S,5R)-1,4,8-trimethyl-2,9-dioxabicyclo[3.3.1]non-7-en-3-yl]hepta-2,4-dien-1-ylidene}-2H-pyrrole-2,4(3H)-dione, PROTOPORPHYRIN IX CONTAINING FE, TamI | | Authors: | Newmister, S.A, Srivastava, K.R, Espinoza, R.V, Haatveit, K.C, Khatri, Y, Martini, R.M, Garcia-Borras, M, Podust, L.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Molecular Basis of Iterative C─H Oxidation by TamI, a Multifunctional P450 monooxygenase from the Tirandamycin Biosynthetic Pathway.
Acs Catalysis, 10, 2020
|
|
6SDR
 
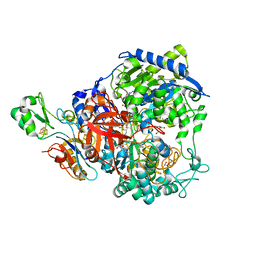 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Oxidized form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, alpha subunit, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
6S8F
 
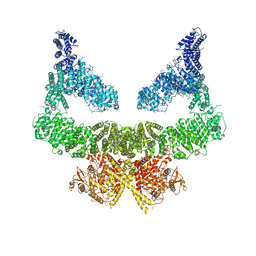 | | Structure of nucleotide-bound Tel1/ATM | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1 | | Authors: | Yates, L.A, Williams, R.M, Ayala, R, Zhang, X. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM Structure of Nucleotide-Bound Tel1ATMUnravels the Molecular Basis of Inhibition and Structural Rationale for Disease-Associated Mutations.
Structure, 28, 2020
|
|
6XRA
 
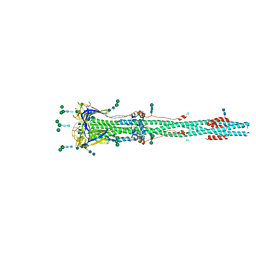 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Rits-Volloch, S, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
5EU7
 
 | | Crystal structure of HIV-1 integrase catalytic core in complex with Fab | | Descriptor: | FAB Heavy Chain, FAB light chain, Integrase | | Authors: | Galilee, M, Griner, S.L, Stroud, R.M, Alian, A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Preserved HTH-Docking Cleft of HIV-1 Integrase Is Functionally Critical.
Structure, 24, 2016
|
|
6XA3
 
 | | Structure of the ligand free P450 monooxygenase TamI | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TamI | | Authors: | Newmister, S.A, Srivastava, K.R, Espinoza, R.V, Haatveit, K.C, Khatri, Y, Martini, R.M, Garcia-Borras, M, Podust, L.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular Basis of Iterative C─H Oxidation by TamI, a Multifunctional P450 monooxygenase from the Tirandamycin Biosynthetic Pathway.
Acs Catalysis, 10, 2020
|
|
7UL6
 
 | |
6SYX
 
 | | Hydrogenase-2 variant R479K - reduced sample exposed to pure oxygen | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
6XTF
 
 | | Crystal structure a Thioredoxin Reductase from Gloeobacter violaceus bound to its electron donor | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Buey, R.M, Gonzalez-Holgado, G, Fernandez-Justel, D, Balsera, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Unexpected diversity of ferredoxin-dependent thioredoxin reductases in cyanobacteria.
Plant Physiol., 186, 2021
|
|
1DTQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTT
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-04-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DVK
 
 | |
